Optimizing image segmentation modeling using Seqera Platform
Scientific research is rarely direct, and workflows commonly require further downstream analyses beyond pipeline runs. While Nextflow excels at batch automation, human interpretation of the generated data is also an essential part of the scientific process. Interactive environments facilitate this process by enabling model refinement and report generation, increasing efficiency and facilitating informed decision-making.
Performing interactive analysis is considered one of the most challenging steps in the entire bioinformatics process. Users face cumbersome, time-consuming, and error-prone manual tasks such as transferring data from the cloud to local storage and navigating various APIs, programming languages, libraries, and tools. User-friendly interactive environments that exist adjacent to your data are critical to streamline end-to-end computational analyses.
Seqera’s Studios bridges the gap between pipeline outputs and secure interactive analysis environments by bringing reproducible, containerized and interactive analytical notebook environments to your data. In this way, the output of one workflow can be analyzed manually and be used as the input for a subsequent workflow. Here, we show how a scientist can use the Seqera Platform’s Runs and Studios features to optimize image segmentation model iteration in the nf-core/molkart pipeline.
Watch the full presentation from Nextflow Summit in Boston, May 2024
How does image segmentation work?
A central task in molecular biology is quantifying the abundance of different molecules (often RNAs or proteins) per cell or structure. Traditionally, this was done by sampling entire tissues or, in later approaches, using single-cell methods to measure such molecules within each cell. However, both bulk and single-cell omics methods lose information about the spatial organization of cells within a tissue, a key factor during tissue development and a potential driver for diseases like cancer. Spatial omics, which combines imaging with ultra-sensitive assays to measure molecules, now allows the identification of hundreds to thousands of transcripts on tissue sections.
nf-core/molkart is a spatial transcriptomics pipeline for processing Molecular Cartography data by Resolve Bioscience, which measures hundreds of RNA transcripts on a tissue section using single-molecule fluorescent in-situ hybridization (smFISH) (Figure 1). This pipeline includes a Nextflow module for the popular segmentation method Cellpose, which allows a human-in-the-loop approach for improving cell segmentation. Conveniently, the nf-core/molkart pipeline includes a workflow branch for generating custom training data from a source data set. Training a performant, custom cellpose model typically requires multiple time consuming human-in-the-loop model iterations within an interactive analysis environment.
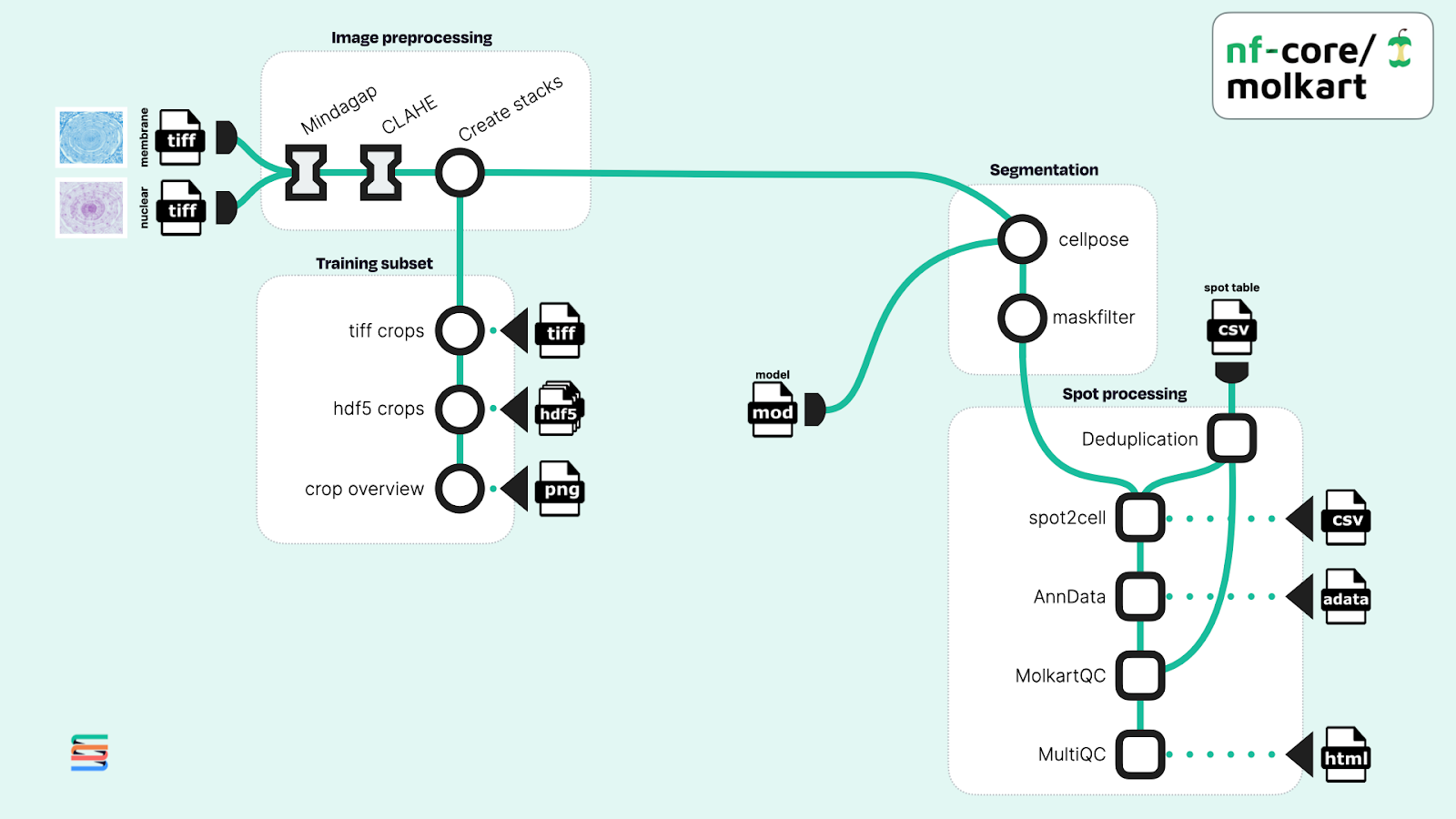
Figure 1. Adapted workflow diagram of the nf-core/molkart pipeline for processing molecular cartography data using Nextflow. Original image data shown was taken from the literature (Perico et al.).
We used Studios to bring the tertiary analysis adjacent to the data in cloud storage, using data from ta 2024 preprint by Perico et. al. This allows us to iteratively train and improve a custom cellpose model for our specific dataset (Figure 2).
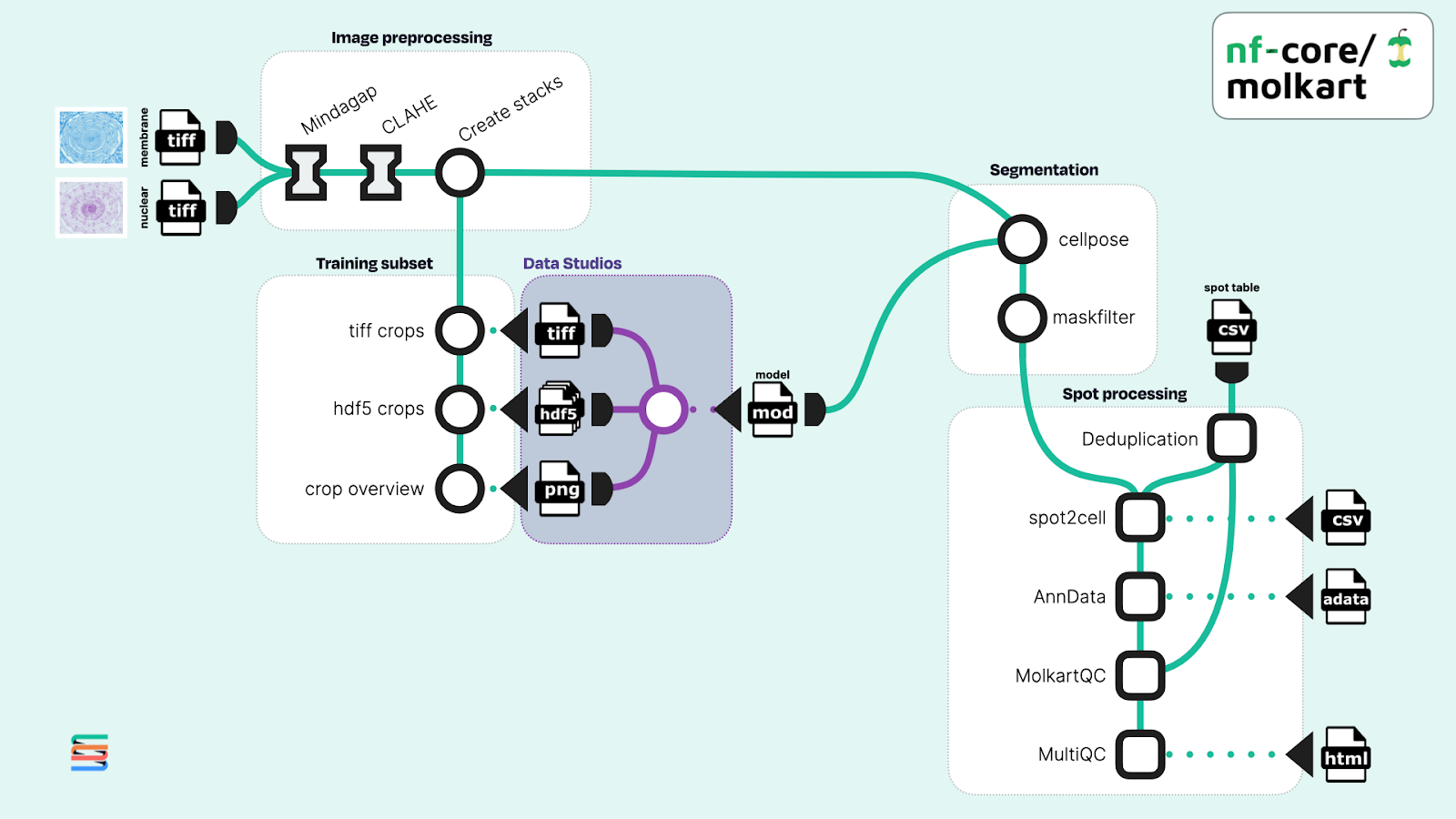
Figure 2. Adapted workflow diagram of the nf-core/molkart pipeline using Studios (highlighted in gray) to iteratively train a custom cellpose model to use as input for cell segmentation. Original image data shown was taken from the literature (Perico et al.).
Adding Studios to the workflow
Using Studios as part of an adapted workflow was extremely beneficial:
- Rapid review of image training data – Images can be quickly reviewed directly in the cloud-hosted Studio analysis environment using common tools such as napari, QuPath, or Fiji. Prior to Studios, bioinformaticians would typically download the images, review, and re-upload to blob storage.
- Collaboratively train a custom model in-situ – Using a GPU-enabled compute environment for the Studios session, we used cellpose to train a new custom model on-the-fly using the previously generated image crops. Using a shareable URL, Studios enables seamless collaboration between data scientists and bench scientists with domain expertise in a single location.
- Apply the new model to the original data – The new, manually-trained model was then applied to the original, full size image dataset. The cell segmentation results of the custom model can be inspected in the same Studios instance using any standard tool.
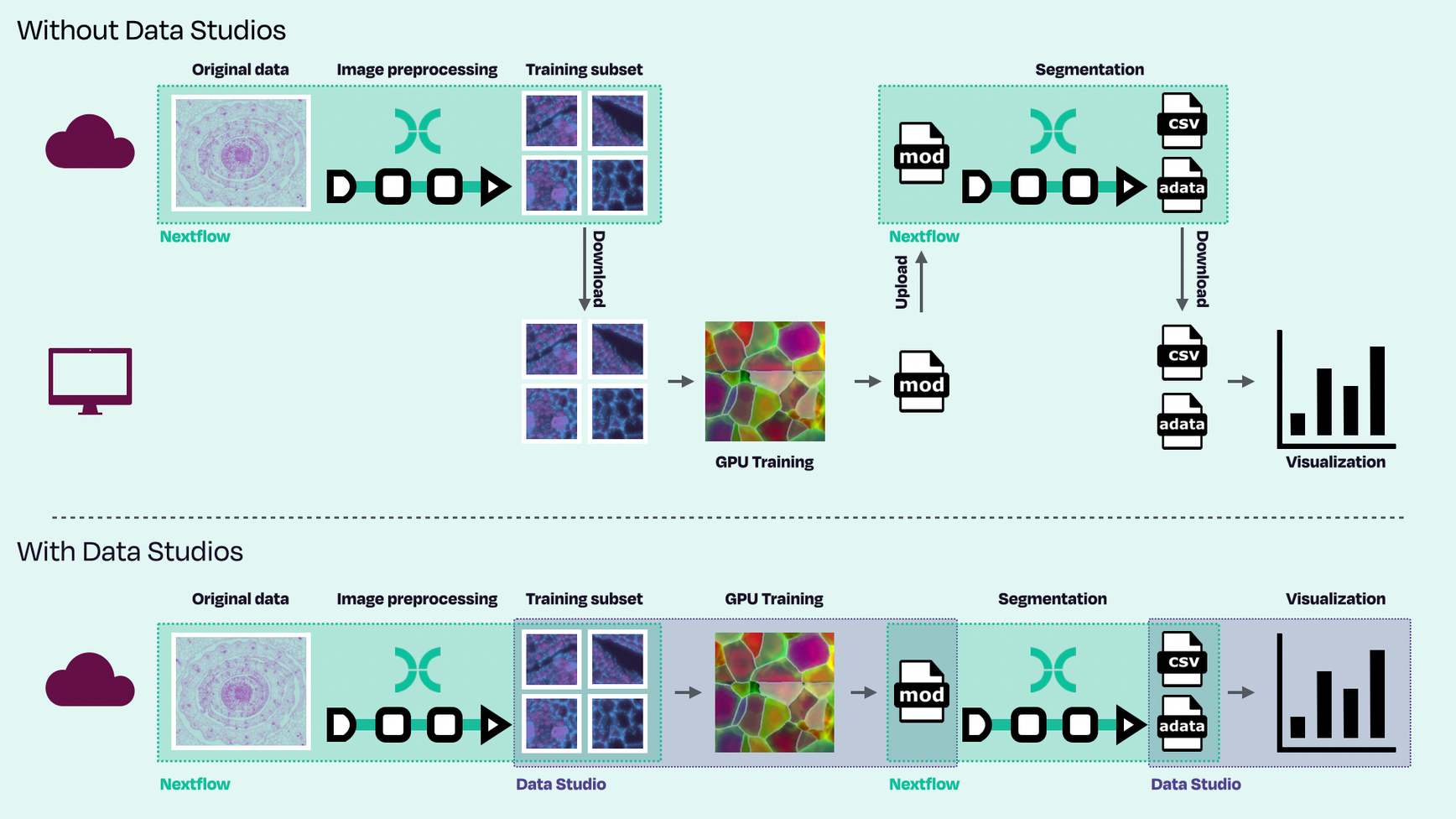
Figure 3. Schematic workflow of image segmentation using nf-core/molkart with (bottom) and without (top) Studios. Original image data shown was taken from the literature (Perico et al.).
The benefits of Studios
- →Data remains in-situ – No shuttling large volumes of data back and forth between your cloud storage and local analysis environments, which can quickly become expensive with ingress and egress charges, is extremely inefficient, and can result in data loss. Using the Fusion file system, Studios enables direct file access to cloud blob storage and is incredibly performant.
- →Stable, containerized analysis environments – Studio sessions are checkpointed, and can be rolled back to any previous state each time the session is stopped and restarted. Each checkpoint preserves the state of the running machine at a point in time, ensuring consistency and reproducibility of the environment, the software used, and data worked with.
- →Provision only the resources you need – Data Studio sessions are fully customizable. Based on the analysis task(s) at hand, they can be provisioned as lean or as fully-featured as required, for example, making them GPU-enabled or adding hundreds of cores.
- →Permissions are centrally managed – Organization and workspace credentials are centrally managed by your organization administrators, ensuring only authenticated users with the appropriate permissions can connect to the data and analysis environment(s). Bioinformaticians and data scientists shouldn’t spend time managing infrastructure and permissions.
- →Secure, real time collaboration – The shareable URL feature ensures safe collaboration within, or across, bioinformatician and data science teams.
Streamline the entire data lifecycle
Studios can streamline the entire end-to-end scientific data lifecycle by bringing reproducible, containerized and interactive analytical notebook environments to your data in real-time. This allows you to seamlessly transition from Nextflow pipeline outputs to secure interactive environments, consolidating data and analytics into one unified location.
“Studios enables the creation of the needed package environment for any project quickly, expediting the project start-up process. This allows us to promptly focus on data analysis and efficiently share the environment with the team”
- Lorena Pantano, PhD
Director of Bioinformatics Platform, Harvard Chan Bioinformatics Core
View Studios in the Seqera Platform Community Showcase workspace or start a free trial today!
