What is AIVA?
AIVA (AI-powered Variant Analysis) is an agentic variant analysis platform that streamlines variant interpretation, clinical reporting, and collaborative genomic analysis. Through natural language queries, AIVA provides an interactive environment for real-time variant interpretation.
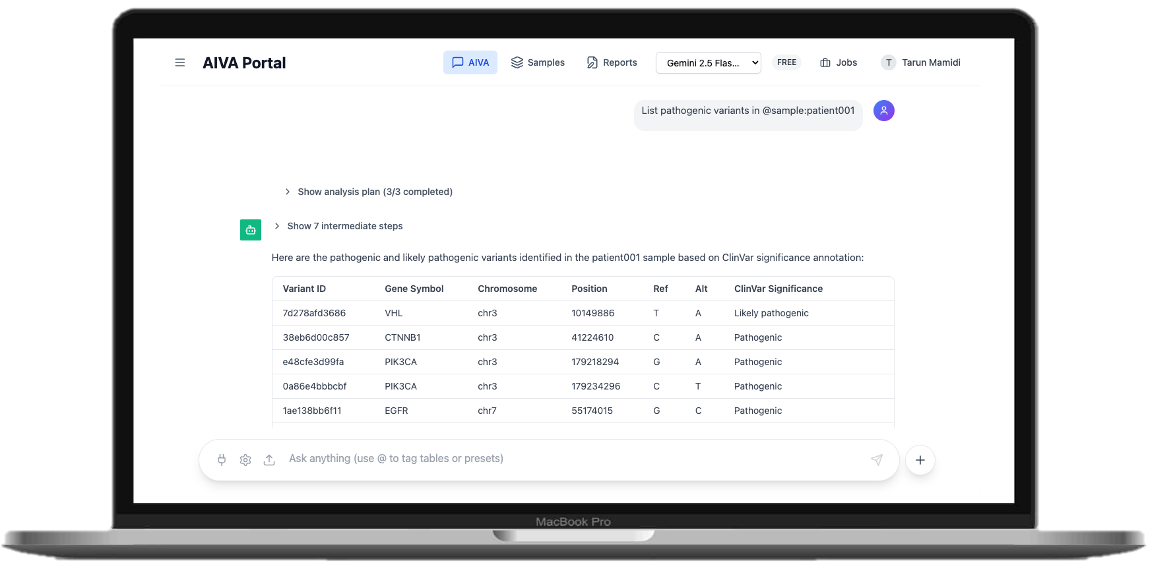
What is Seqera Studios?
Studios, a powerful feature of Seqera, streamlines the end-to-end data lifecycle by bringing reproducible, fully customizable interactive analytical environments to your data in real-time. With Studios, you can seamlessly transition from pipeline results to secure interactive analysis environments for immediate exploration, without moving data or leaving your cloud.
Key Benefits of Seqera Studios
- →One-click data access: Mount cloud storage directly in your workspace; interact with data as if it were local.
- →Reproducible environments: Every Studio is a tagged container image, ensuring consistent, reproducible setups that can be reopened or shared anytime.
- →Fully customizable: Use pre-built templates, modify existing ones, or bring your own Docker images and application stacks.
- →Real-time collaboration: Share sessions via URL for simultaneous multi-user access to notebooks and dashboards.
How and why AIVA decided to integrate with Studios?
One of the key challenges in genomics analysis is the disconnect between platforms that offer secondary analysis pipelines and variant interpretation. While most of these secondary analysis pipelines can be customized, analyzing variants is limited to external platforms which need moving data between platforms. AIVA brings AI variant analysis closer to secondary variant calling pipelines to improve workflow efficiency.
- →Eliminating Data Gravity: Performing tertiary analysis within Studios, next to secondary analysis pipeline output, avoids costly and time-consuming data transfers.
- →Seamless Workflow Transition: Studios acts as a bridge from secondary analysis (via Seqera Platform) to interactive, AI-powered tertiary interpretation and reporting with AIVA.
This seamless integration allows you to transition between your customized bioinformatics pipelines and genomic analysis effortlessly, all within Seqera.
Launch AIVA in Seqera Studios
Follow these simple steps to integrate AIVA into your Platform workflow:
Prerequisites
• VCFs from any secondary analysis pipeline (e.g., nf-core/sarek, raredisease)
• Access to Seqera Platform and compute environment
• AIVA Portal account at chat.aivaportal.com
💡 Important: AIVA does allow you to bring your own annotations. Please use VEP/SnpEff/BCFTools or from your Sarek/raredisease pipelines to bring your own annotations.
Step-by-Step Setup
- Navigate to Seqera and run your variant calling pipeline (Sarek, raredisease, etc.)
- Use Data Explorer to locate your VCF file in your cloud bucket and copy the URL path
- Click "Studios" → "Add Studio" to create a new Studio session
- Click "Mount data" and select the bucket containing your VCF files
- Configure the Studio with AIVA's prebuilt container image:
- Change from template to "Prebuilt container image"
- Container identifier: gcr.io/aiva-b2387/aiva-chat-seqera:latest
- Click "Add and start"
- Wait for Studio to spin up the AIVA instance (30-60 seconds)
Once ready, click "Login" to access the AIVA Portal
How to Use AIVA
Import Your VCF Files
After logging into the AIVA Portal, you'll need to import your VCF files from the mounted cloud storage.
Step-by-Step Guide
- →Navigate to "Samples" in the AIVA Portal
- →Click "Upload Files" to access the file import options
- →Optional - select “Enable variant annotation” if you haven’t annotated your VCF in the secondary analysis pipeline.
- →Select "Cloud Import" tab
- →Paste your VCF URL in the import field
- →Replace
s3://orgs://with/workspace/data/since the bucket is already mounted. - →Example:
/workspace/data/your-bucket-name/sample1-annotated.vcf.gz
- →Replace
- →Click "Import from Cloud" to start the import process
- →Click "Jobs" to monitor the upload progress (larger files may take longer to process)
- →Once processing is complete, click "Browse by Samples" to find your uploaded samples
Start AI-Powered Analysis
Now you can begin analyzing your genomic data using AIVA's AI assistant.
- →Click "Chat" to open the AI analysis interface
- →Start analyzing based on the annotations in your VCF file using natural language queries
- →AIVA's AI agent executes steps transparently and returns comprehensive responses
Example Queries:
• "Flag all pathogenic variants in BRCA1 with MAF < 0.01 in @patient01"
• "Find genes associated with Thyroid cancer and Show me loss-of-function variants in @patient01"
• "Classify BRAF V600E using ACMG criteria for Colorectal Cancer
• "Generate a variant type distribution plot for all flagged variants"
With AIVA, you can perform variant interpretation by just simple conversation, leveraging the power of state-of-the-art GenAI models. Explore variants, classify using ACMG/AMP criteria, flag them and generate a report all within the same chat window. Integrating with Studios keeps your whole workflow in the same platform to perform end-to-end variant analysis.
Interested in learning more? Check out the resources below.
• Learn more about AIVA
• Discover Seqera Studios
About Mamidi Health

Mamidi Health is advancing genetic disease diagnosis through autonomous clinical workflow agents that accelerate the identification of disease-causing genetic variants, reducing turnaround time while improving affordability and diagnostic confidence. Visit https://mamidi.ai/ for more information

