Last month, the developers of AlphaFold2 were awarded the 2024 Nobel Prize in Chemistry. Did you know you can easily perform 3D protein structure prediction analysis using AlphaFold2 and other AI models in Seqera?
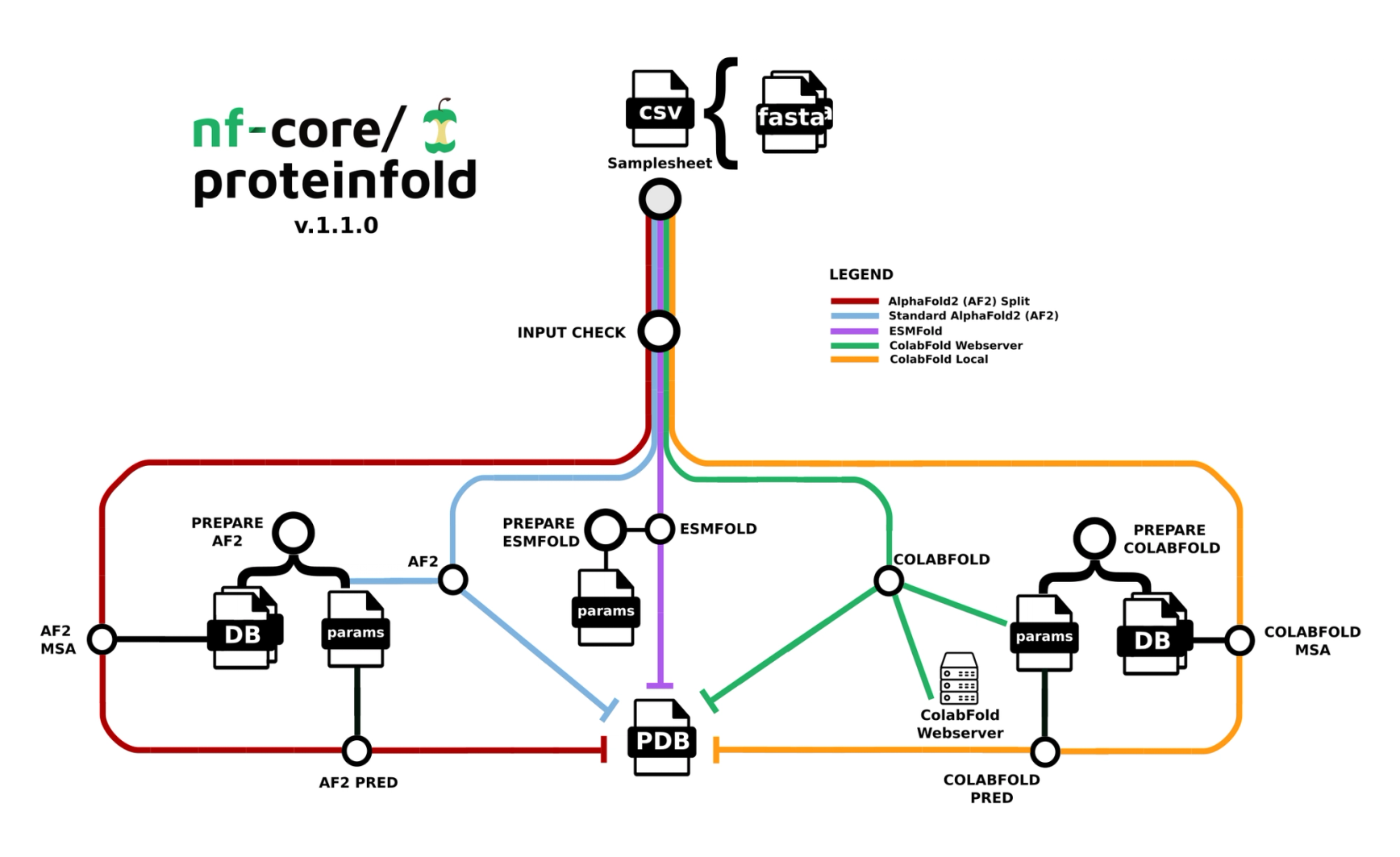
We are excited to present the next blog post in our Step-by-Step series on running Nextflow community pipelines in Seqera. In this installment, we show how to perform and interactively visualize 3D protein structure predictions with the nf-core/proteinfold pipeline and Studios in Seqera Platform.
1. Add a compute environment
Create a Platform compute environment that meets the data and computational demands of protein folding workflows:
✔ GPUs for optimal performance.
✔ Multi-core CPUs for non-GPU tasks.
✔ Low-latency, high-throughput Fusion file system.
💡Hint: The compute and storage requirements for protein structure prediction depend on the number and length of protein sequences being analyzed and the size of the database used for prediction by the deep learning models.
Check out the full protein structure prediction guide now
2. Add nf-core/proteinfold to your workspace
Use Seqera Pipelines to quickly add the nf-core/proteinfold pipeline to your Platform workspace.
Seqera Pipelines is the largest curated open source repository of Nextflow pipelines.
3. Add your data
Access your reference and raw sequence data directly from your Platform workspace:
- →Browse and interact with remote cloud storage data using Data Explorer.
- →Upload structured data as CSV or TSV files using Datasets.
4. Launch your protein prediction analysis
With your compute environment, nf-core/proteinfold pipeline, and experimental and reference data all accessible in Seqera, you are ready to launch your protein prediction analysis.
💡Hint: Select which deep learning model to use for structure prediction (AlphaFold2, ColabFold, or ESMFold) from the “Mode” menu.
5. Create a flexible interactive analysis environment
Use built-in templates to easily create a custom environment and add the packages, libraries, and scripts required for your analysis.
💡Hint: Studios Jupyter, R IDE, VSCode, and Xpra environments are infinitely customizable to your analysis or troubleshooting needs.
6. Visualize and compare predicted protein structures
Protein structure prediction often requires further analysis, such as structure visualization.
Use tools such as Biopython and NGLView to create an interactive visualization comparing different models’ predictions in your preferred notebook environment.
Here we use a Jupyter notebook to compare AlphaFold2 and ColabFold 3D protein structures.
7. Perform collaborative analysis in real time
Share your data studio session URL to collaborate with others and cross-validate findings in real time.
Any workspace users with the Connect role can join your data studio sessions for real-time collaboration.
Want to learn more?
By leveraging cloud-native technology, Seqera bridges the gap between experimental data and computational analysis, allowing you to accelerate the time from data generation to meaningful scientific insights.
See the full guide to start your protein folding analysis now.
