Seqera Integrates NVIDIA Parabricks and Fusion to Seamlessly Accelerate Genomics Analysis
As data volumes explode in the life sciences, computational bottlenecks threaten to slow scientific discovery. To address this, Seqera has integrated NVIDIA Parabricks, a GPU-accelerated software suite for genomic analysis, and Fusion, Seqera’s optimized filesystem for cloud-native Nextflow pipelines. Together, these technologies dramatically accelerate data analysis, reduce costs, and simplify cloud infrastructure for bioinformatics teams.
Learn more about FusionNVIDIA Parabricks: Accelerating Data Analysis with AI and GPUs
NVIDIA Parabricks is an AI-accelerated, GPU-based software suite designed for high-performance omics analysis. It delivers dramatic acceleration across secondary analysis tasks such as multi-omics alignment—with tools like BWA-MEM, minimap2, and STAR—and high-accuracy variant calling using tools including DeepVariant and HaplotypeCaller. Parabricks is distributed via Docker containers and is already well-integrated into the nf-core ecosystem, with nine modules readily available for use. In addition to Parabricks, NVIDIA also supports tertiary analysis through tools like Evo2 and Geneformer in BioNeMo Framework.
💡nf-core Parabricks modules currently available: Parabricks_haplotypecaller, Parabricks_deepvariant, Parabricks_mutectcaller, Parabricks_genotypegvcf, Parabricks_indexgvcf, Parabricks_fq2bammeth, Parabricks_fq2bam, Parabricks_applybqsr, Parabricks_dbsnpLearn more about NVIDIA Parabricks
Key Benefits of NVIDIA Parabricks
One key advantage of Parabricks is speed. Benchmarking by NVIDIA showed 135x faster analysis of WGS data compared to CPU-only, reducing germline analysis time from 18 hours to just 8 minutes with RTX PRO 6000 Blackwell Server Edition. In addition to performance gains, Parabricks significantly lowers costs—achieving up to 50% reduction in compute expenses for WGS compared to CPUs—and enhances analytical precision through deep learning-powered, high-accuracy variant calling.
Fusion: Removing Storage Bottlenecks in Data-intensive Workflows
Modern genomics workflows are often limited not only by processing power, but also by data throughput and I/O performance. Traditional POSIX-based file systems (whether shared or distributed) struggle to handle the growing scale of sequencing data. These legacy systems require costly infrastructure, manual data staging from cloud storage, and slow pipeline execution due to data movement overhead.
Fusion enables direct, transparent access to cloud object storage—eliminating the need to stage files locally. This allows pipelines to analyze data in place, accelerating execution while simplifying infrastructure. Fusion removes traditional I/O bottlenecks, supports elastic storage scaling (with no need to pre-provision disks), and offers a highly cost-effective model where you pay only for what you use. For large-scale genomics, Fusion makes cloud-native processing faster, cheaper, and significantly easier to manage.
Key Benefits of Fusion
Fusion fully optimizes how Nextflow interacts with data by enabling direct access to cloud object storage. This results in breakthrough pipeline performance, delivering up to 2.2× greater throughput compared to standard S3 access. It also drives significant cost efficiency, with up to 76% reductions in pipeline and storage expenses. By minimizing data movement, Fusion enhances portability and reliability, making workflows more robust, reproducible, and easier to scale across diverse cloud environments.
Interested in learning more about Fusion? Read our latest case study on streamlining bioinformatics at Bio-Rad with Fusion for faster workflows, cost-efficiency, and simplified infrastructure.
Read the Fusion Case StudyThe Power of Integration: NVIDIA Parabricks and Fusion
The integration of Parabricks and Fusion brings together two breakthrough technologies that dramatically improve the speed and efficiency of omics workflows:
- →Parabricks accelerates compute-intensive steps with high-throughput, GPU-based processing for dramatically faster runtimes
- →Fusion eliminates I/O bottlenecks with seamless, cloud-native data access and optimized storage-to-compute performance.
Together, they deliver end-to-end workflow acceleration, combining fast compute with seamless, high-throughput data access. This helps researchers process samples in less time, avoid expensive storage overprovisioning, and accelerate time-to-insight without compromising accuracy.
Benchmarking with nf-core/sarek
To demonstrate real-world impact, the nf-core/sarek pipeline for germline variant calling was benchmarked on 30X WGS data and compared against two setups: (i) CPU-only execution and (ii) Parabricks without Fusion. For this benchmarking, we modified key compute-intensive steps in nf-core/sarek to leverage GPU acceleration:
- →Replaced BWA and GATK MarkDuplicates with Parabricks fq2bam
- →Implemented Parabricks DeepVariant for variant calling
10x Faster Pipeline Runtime
The standard nf-core/sarek pipeline (BWA and GATK) running on CPUs served as the baseline, taking almost 20 hours to run. Switching to Parabricks on GPUs achieved a 7.7x speedup, and combining with Fusion pushed performance further to a 10x overall acceleration at just under 2 hours.
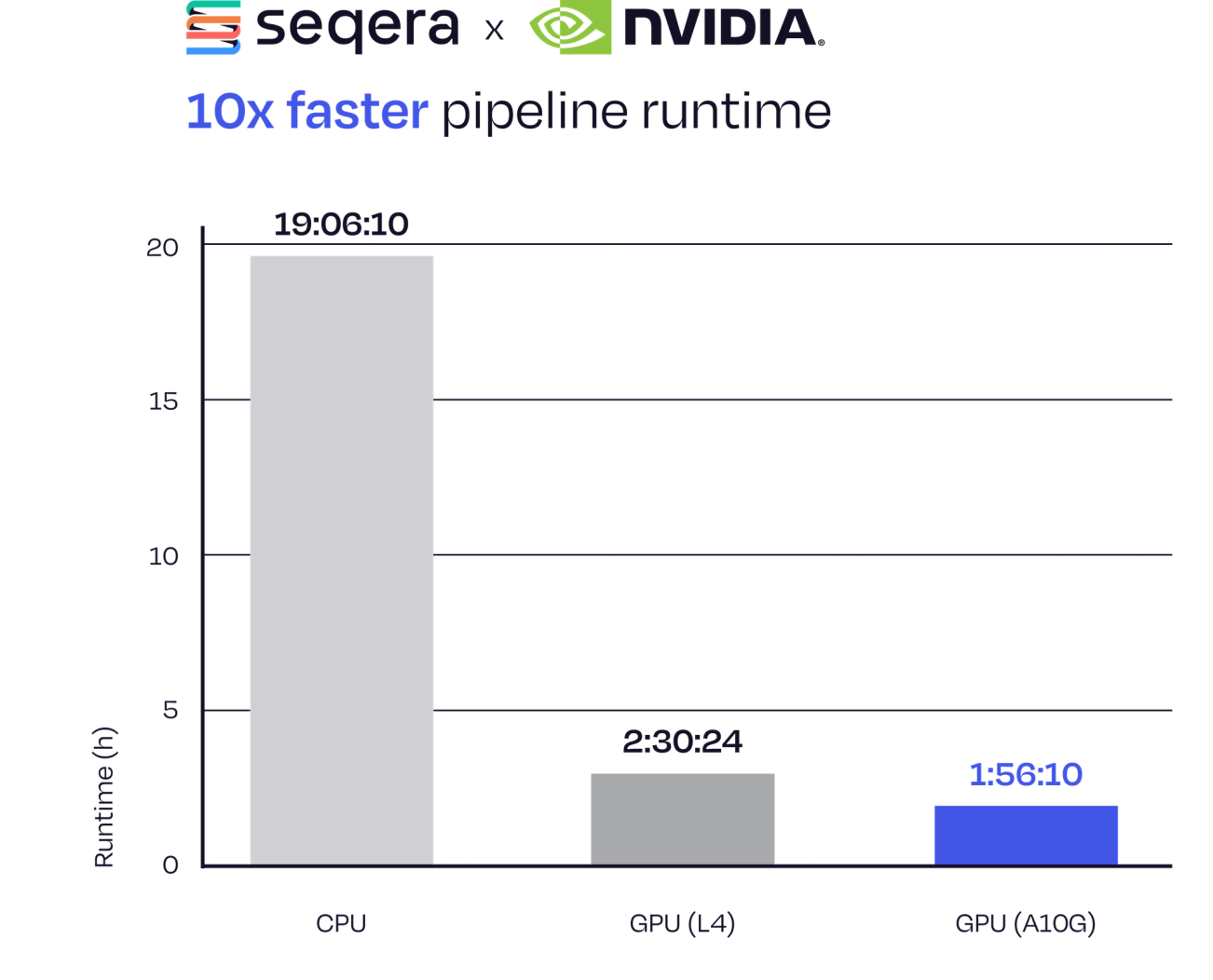
Figure 1. 10x speedup in nf-core/sarek using the Parabricks and Fusion integration.
65% Reduction in Compute Cost
The integration of Parabricks and Fusion delivered significant cost-savings. Compared to the standard pipeline run on CPUs, using Parabricks alone reduced compute costs by ~38%. Combining this with Fusion resulted in an overall 65% reduction in compute costs.
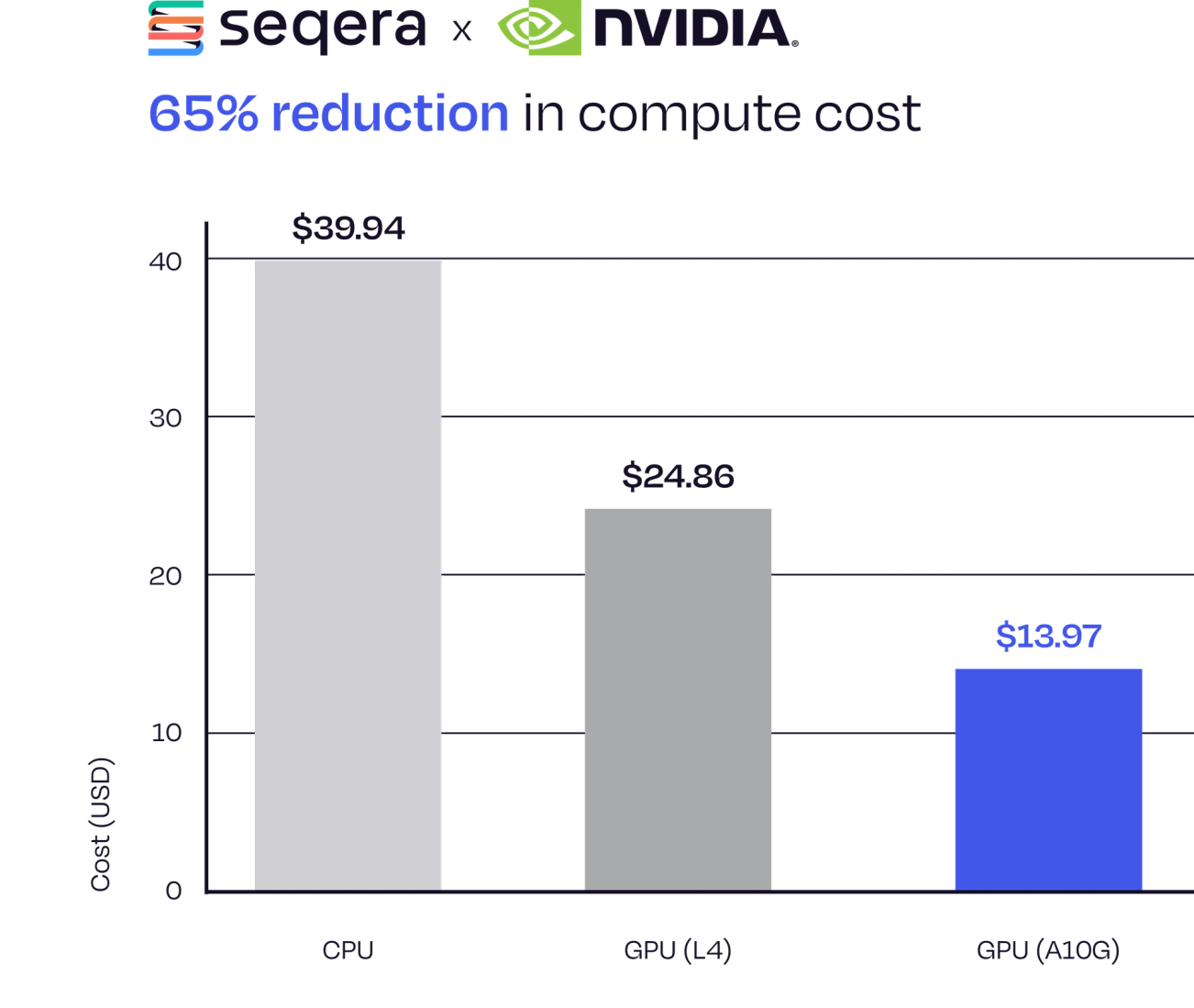
Figure 2. 65% reduction in compute cost for running nf-core/sarek using the Parabricks and Fusion integration.
Further 57% Cost Reduction with Spot Instances
Fusion enables the optimal use of spot instances in your Nextflow pipelines, with zero-cost restarts. We have already shown compute cost-savings of 65% using the Parabricks Fusion integration compared to on-demand CPU runs (Figure 2). Leveraging spot instances further reduces costs by 57% (from $13.97 to $6.07), maximizing efficiency without compromising reliability.
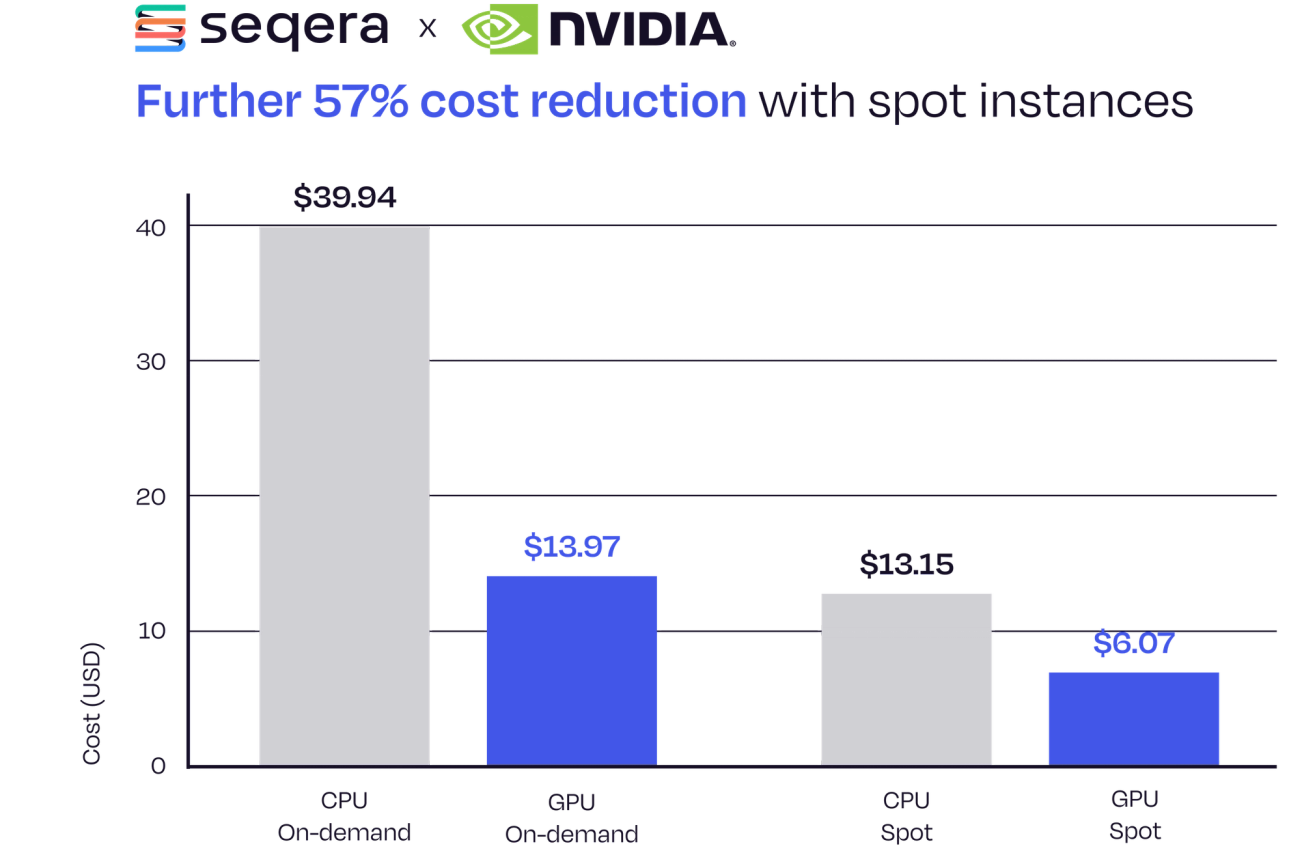
Figure 3. Further 57% reduction in compute cost for running nf-core/sarek using the Parabricks and Fusion integration with spot instances compared to on-demand.
Summary of Benchmarking Results
These results highlight the practical benefits of integrating Parabricks and Fusion in compute-intensive Nextflow pipelines, delivering faster runtimes and significant compute cost-savings over traditional CPU-based execution. By accelerating key processing steps and simplifying data access, this approach helps make high-throughput genomics analysis more efficient, affordable, and easier to scale in cloud environments.
A notable benefit is the flexibility to dynamically allocate resources by seamlessly switching between CPU and GPU instances based on performance and budget needs. This integration also enables Parabricks to be embedded as an optional component within existing workflows, delivering speed-ups for compute-heavy tasks while allowing interoperability with other widely used toolkits, such as those for quality control.
Future work
There remains considerable potential for further optimization. Leveraging single-interval variant calling can deliver additional speedups by enabling more granular parallelism. Fusion helps eliminate staging bottlenecks, particularly for large genomic datasets, by providing direct, scalable access to data without unnecessary duplication. Additionally, auto-scaling storage dynamically adjusts resource allocation, minimizing over-provisioning and reducing cloud infrastructure costs.
Seamless Deployment in Seqera Platform
Deploying this integration in Seqera Platform is straightforward and requires minimal changes to your existing Nextflow workflows. Getting started is simple: when configuring your workflow, just select “Use Amazon-recommended GPU-optimized ECS AMI” to enable GPU acceleration with Fusion and Parabricks when setting up your compute environment. Seqera Platform automatically manages complex infrastructure, eliminating the typical hurdles of getting started with GPUs at scale.
 Try the Integration for yourself
Try the Integration for yourselfAcknowledgements and Community Contributions
These optimizations are being actively contributed back to the nf-core community, ensuring broader access and long-term sustainability. The Parabricks integration is designed as an alternative execution path within existing pipelines, preserving compatibility and minimizing disruption. There’s no need to rewrite your workflows, simply leverage familiar nf-core structures while benefiting from GPU acceleration and Fusion optimizations.
💡Interested in learning more?
• Find Parabricks modules on nf-core
• Discover the benefits of Fusion for your workflows
• Log in to Seqera to try the integration yourself



