Nextflow and Tower for Machine Learning
“Nextflow” and “machine learning” are not topics that are typically seen together. Nextflow is usually associated with bioinformatics, while machine learning has been used mostly for computer vision, speech recognition, and natural language processing.
In reality, however, there’s no reason why you can’t use Nextflow for machine learning (or machine learning for Nextflow!). Nextflow is a general-purpose workflow language; it can use a machine learning model just like any other software tool. And machine learning is increasingly being applied to every domain – including bioinformatics. According to Data Bridge Market Research, AI in bioinformatics is poised to grow by 42.7% on average each year through 2029.
So how can you create machine learning pipelines with Nextflow? In this article, we’ll show you just that. In particular, we’ll discuss the following:
- Examples of machine learning applications for life sciences and healthcare
- A simple Nextflow pipeline for training and evaluating an ML model
- Model deployment with Nextflow and Tower
By the way, if you’re just getting started with machine learning, there are plenty of excellent machine learning primers that can introduce the topic much better than we could here. Here are a few resources to get you started:
- DeepLearning.AI: Machine Learning Specialization
- Kaggle: Intro to Machine Learning
- TensorFlow: Basics of machine learning
Machine learning in life sciences and healthcare
Machine learning is already being used for a wide variety of applications in life sciences and health care. Here are just a few examples:
- Medical imaging — scan medical images for various types of anomalies1.
- Biomarker discovery — identify specific genes (i.e. “biomarkers”) that are associated with the development of specific cancers, using RNA sequencing data from healthy and tumorous tissue samples.2
- Protein folding — predict the 3D structure of proteins from their amino acid sequences.3
- Drug discovery — model and predict how various drug compounds interact with target proteins or with one another, e.g. to find effective drugs for treating specific diseases.4
- Gene editing — facilitating gene editing experiments with CRISPR by improving accuracy in selecting the correct DNA sequences for manipulation.5
Machine learning itself encompasses a broad range of techniques, from basic statistical methods like linear regression, to “classic” machine learning algorithms such as k-nearest neighbors and random forests, to “deep learning” techniques such as convolutional neural networks and transformers.
For bioinformaticians looking to apply machine learning approaches to their domain, the challenge lies in selecting from this wide range of techniques, and finding a model that achieves sufficient “performance” with their data. As with any machine learning application, “performance” could incorporate any number of metrics, such as accuracy, sensitivity / specificity, and compute cost. Once a sufficiently performant model is found, it can be deployed like any other software application to perform inference on new data.
To illustrate how Nextflow and Tower can help with both model training and inference, let’s focus on a specific example.
Case study: Breast Cancer
The Wisconsin Diagnostic Breast Cancer (WDBC) dataset is a well-known dataset that is often used as an example for learning or comparing different machine learning techniques, specifically for classification. The dataset consists of 589 samples, and each sample is a set of 30 features taken from an image of a breast tissue. The diagnosis column indicates whether the sample was benign (B) or malignant (M), as illustrated below.
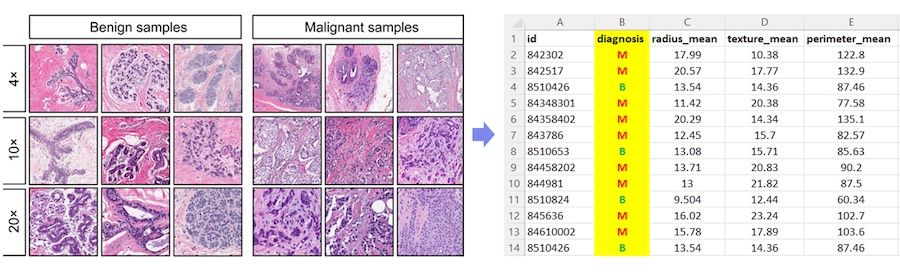
Image from Breast Cancer Wisconsin (Diagnostic) Dataset - Exploratory Data Analysis by Shubam Sumbria
For our case study, we will train and evaluate a variety of models on the task of classifying these breast samples as benign or malignant. In a real-world scenario, we would probably use k-fold cross-validation to evaluate each model on several randomized partitions of the dataset, and use several performance metrics with minimum requirements to determine whether a model is “good enough” to be used in the wild. For the purposes of this example, we will simply evaluate each model on a single 80/20 train/test split and select the model with the highest test accuracy.
Creating a machine learning pipeline with Nextflow
We developed a proof-of-concept pipeline called hyperopt to demonstrate a model training pipeline in Nextflow. The pipeline takes any tabular dataset (or the name of a dataset on OpenML), and trains and evaluates a set of machine learning models on the dataset, reporting the model that achieved the highest test accuracy.
The pipeline code is available on GitHub. Here is a Mermaid diagram of the overall workflow:
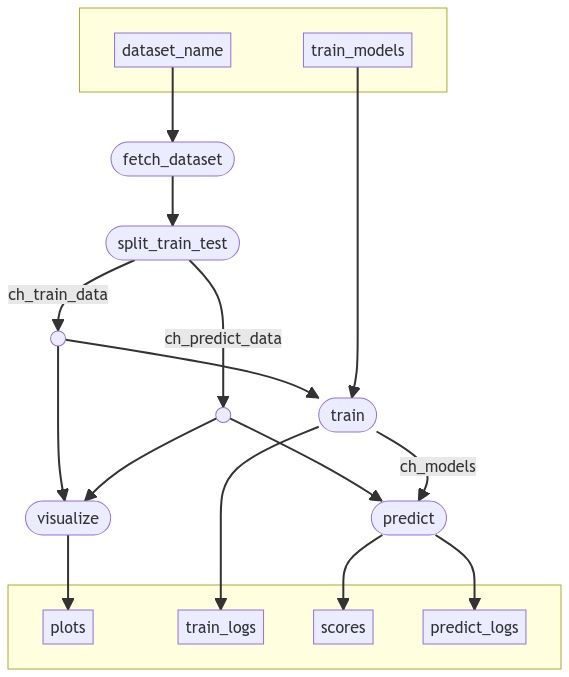
The pipeline steps are implemented as Python scripts that use several common packages for machine learning, including numpy, pandas, scikit-learn, and matplotlib. These dependencies are defined in a Conda environment file called conda.yml.
There are two ways to run the pipeline – the Conda approach and the Wave approach.
- If you run the pipeline with
-profile conda, Nextflow will create a Conda environment with the Python dependencies (including Python itself) and cache it for subsequent runs. This approach requires Conda to be installed in your launch environment. - If you run the pipeline with
-profile wave, Nextflow will use the Wave container service to build a Docker image with the Python dependencies. This approach requires Docker to be installed in your launch environment.
These approaches make it easy to use Nextflow with Python scripts. You need only to define your Python dependencies in a Conda environment file, and Nextflow will handle the rest. Refer to the Nextflow documentation on Conda and Wave to learn how to configure them further.
By default, the pipeline uses the aforementioned WDBC dataset and evaluates five different classification models:
- A baseline model (i.e. “dummy” model) that simply predicts the most common label
- Gradient Boosting
- Logistic Regression
- Multi-layer Perceptron (i.e. neural network)
- Random Forest
When you run the pipeline, you should see something like this:
While the hyperopt pipeline is very basic, it provides all of the building blocks you need to create your own machine learning pipelines with Nextflow. Here are just a few ways that you could extend this pipeline for more advanced use cases:
- User-defined models — the pipeline supports a fixed set of models from scikit-learn, but could be extended to support user-defined models based on any framework, including scikit-learn, TensorFlow, or PyTorch.
- More hyperparameters — despite the name “hyperopt”, which is short for “hyperparameter optimization”, the pipeline has only one hyperparameter, that is, which model to use! This pipeline could be expanded to also search the specific hyperparameters of each model, rather than simply using the default settings.
- Unstructured datasets — the pipeline supports tabular datasets, but could be extended to support other types of data such as images, audio, 3D models, and free-form text.
- Pre-processing, cross-validation, post-processing, performance metrics, etc — the list goes on, but there are numerous components of real-world machine learning pipelines that are not included in this example pipeline in order to keep it simple and generic.
Feel free to use the hyperopt pipeline as a starting point for your next machine learning project!
Scaling model training and inference with Tower
When it comes to training and evaluating machine learning models, Nextflow offers several capabilities that make it ideal:
- Nextflow’s dataflow programming model makes it easy to build and maintain a machine learning pipeline as it grows in size and complexity.
- Nextflow’s integration with many different execution environments makes it easy to take your machine learning pipeline from small experimentation on your laptop or HPC cluster to real-time inference in the cloud.
- Nextflow’s integration with various Git providers and container registries makes it easy to run specific versions of your code in a reproducible manner, in an environment where datasets and algorithms are constantly evolving.
Additionally, Tower is extremely beneficial for collaboration, data management, and model deployment:
- Researchers can collaborate on machine learning models in workspaces, where they can share pipelines, datasets, and results.
- Users can leverage Tower Datasets to organize versioned, labeled training data to simplify management and ensure repeatability.
- With Tower Forge and built-in support for spot instances and GPUs, researchers can run compute-intensive model-training workloads in a cost-efficient manner.
- Tower Pipeline Actions can be used to deploy a model for real-time inference, or retrain a model as new data becomes available.
You can run the hyperopt example pipeline with Tower by adding it as a Pipeline and running it with the wave profile on a Wave-enabled AWS Batch compute environment. For a more complete example, see our previous blog post in which we deploy a Stable Diffusion model as a Nextflow pipeline: Running AI workloads in the cloud with Nextflow Tower—a step-by-step guide.
Nextflow pipelines already using machine learning
As a matter of fact, researchers and developers are already creating machine learning pipelines with Nextflow!
EASI-FISH (Expansion-Assisted Iterative Fluorescence In Situ Hybridization), developed at the Janelia Research Campus, is a Nextflow pipeline that uses image analysis to study the brain. EASI-FISH processes very large image sets (~10TB per experiment), surveys gene expressions in brain tissue, and embeds a deep-learning based cell segmentation model. The pipeline code is publicly available on GitHub.
Nextflow is also used for brain diffusion MRI image workflows by Imeka, a biotech company located in Sherbrooke, Quebec. By combining advanced white matter imaging with artificial intelligence, Imeka’s ANIE (Advanced Neuro Imaging Endpoints) biomarker platform analyzes white matter pathology in greater detail than anyone else. Read our case study for more information about how Imeka uses Nextflow to train high-quality predictive models.
I myself contributed to a few machine learning pipelines in Nextflow during my time in graduate school. TSPG, short for Transcriptome State Perturbation Generator, uses a generative adversarial network (GAN) trained on gene expression data to identify genes that were closely associated with the transition from healthy tissue to cancerous tissue, even for an individual cancer patient. Both the paper and code are publicly available.
View Konrad Rokicki’s talk on Large scale image processing with Nextflow from the 2022 Nextflow summit.
Learn more
The hyperopt pipeline provides an excellent starting point for experimenting with machine learning pipelines in Nextflow. Feel free to fork the pipeline, make changes, and experiment with it in Tower Cloud, which you can access with a free account.
If you are new to Nextflow and Tower, we’ve assembled an excellent collection of learning resources in this article: Learn Nextflow in 2023.
1 Imeka ⎯ a use case involving Nextflow for AI-based processing of diffusion MRI images using PyTorch
2 Cellular State Transformations Using Deep Learning for Precision Medicine Applications
3 Highly accurate protein structure prediction with AlphaFold
4 ORNL team enlists world’s fastest supercomputer to combat the coronavirus