The Data Lifecycle in 2025
In the early days of omics research, generating data was the primary bottleneck, constrained by high costs, low-throughput sequencing, and limitations in automation. Today, sequencing and imaging is faster, cheaper, and more accessible than ever. Research is no longer constrained by what we can sequence, but by what we can meaningfully deduce, forcing a paradigm shift in how we approach computation, storage, and interpretation. Advances in bioinformatics, better integration with data repositories, and new omics technologies are crucial for enabling researchers to realize the full potential of their data.
This evolving landscape is fueling data-driven innovation which is revolutionizing drug development, reshaping how scientists generate, access, and analyze information at scale. From leveraging deep learning algorithms to predict protein structures to using AI models for cancer detection, these innovations are enabling faster, more accurate insights. Such innovations are making a profound impact in areas like disease understanding, drug discovery, and personalized medicine, driving breakthroughs and accelerating the development of targeted therapies.
Yet, despite these advancements, the scientific data lifecycle remains far from seamless: laboratories remain disconnected, data is often inaccessible, and analyses are fragmented. These challenges make it difficult to fully harness the potential of data. In this blog post, we’ll explore these challenges and highlight key considerations for each stage of the modern data lifecycle, ultimately driving greater efficiency and enabling more impactful scientific discoveries.
Key Considerations for the Scientific Data Lifecycle
The modern data lifecycle spans from data generation to analysis and application. Each stage presents unique challenges that can slow progress and limit the full potential of scientific data. Here, we outline key strategies to improve and streamline the data lifecycle, helping to eliminate analytics bottlenecks.
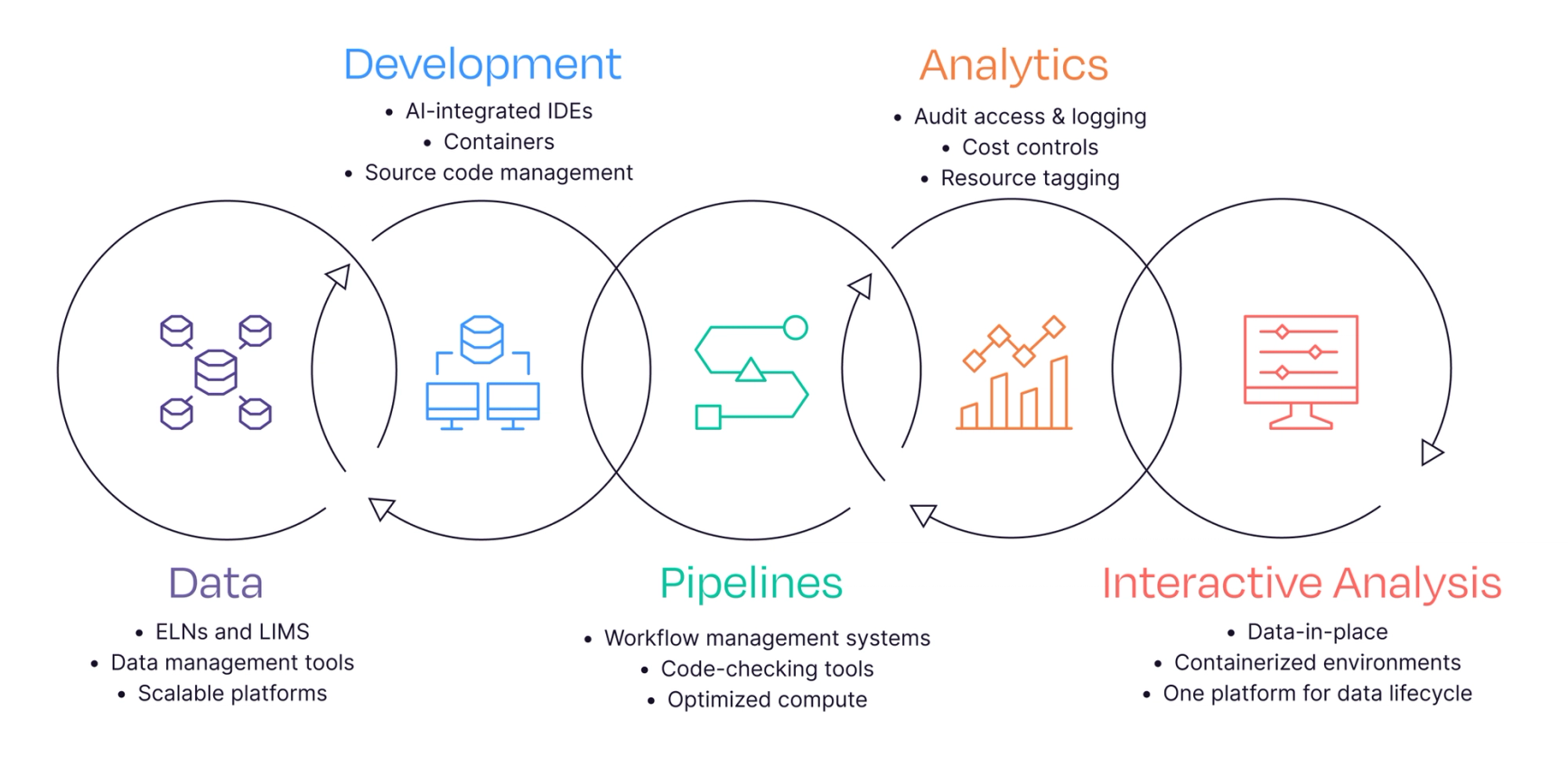
1. Data Management and Generation
The generation of data is the first critical step in the scientific data lifecycle. In omics research, the challenge lies in the sheer volume of data being generated and enriching that data with the relevant experimental metadata. Additionally, data often comes from disparate sources, is poorly annotated, and exists in different formats. These challenges are further compounded when experiments are conducted across multiple labs and organizations, creating substantial infrastructure overheads for secure storage, management, and sharing of data. Scientists need to move towards version-controlled data structures, in similar ways Git is used for code.
💡Consideration: Consistent Data Collection and Management
✔ Link data to experiments with ELNs (e.g., Benchling) track with LIMS.
✔ Catalog data and metadata with data management tools (e.g., TileDB, Quilt, and Lamin).
✔ Standardize data collection protocols across teams and organizations.
✔ Use scalable cloud-native and on-premises platforms.
2. Development
For years, data scientists have spent 80% of their efforts finding and preparing data, and only 20% on actual analytics, underscoring the need for more efficient development environments. The development phase of the data lifecycle centers on designing and building tools, workflows, and infrastructure to standardize and transform data efficiently. However, selecting the right tools presents a challenge, as solutions must balance scalability, reproducibility, and integration across diverse environments. As a result, researchers often use AI-integrated IDEs to develop within containerized, production-like environments, as well as reproducible, cloud development environments like GitHub Codespaces to ensure consistency and scalability. AI-assisted IDEs and reproducible dev environments mirror best practices in software engineering. Additionally, automated workflows, custom tools, and scalable computational environments help manage dependencies, ensure compatibility across tools, and optimize workflows for large-scale data processing.
💡Consideration: Scalable Tools and Containerized Environments
✔ Leverage AI-integrated IDEs such as the Nextflow VS Code extension, Seqera AI, Cursor, or Windsurf).
✔ Utilize containers in development (e.g. using Docker or Apptainer).
✔ Use source code management platforms like GitHub.
3. Pipelines
Data analysis pipelines are an effective way to execute bioinformatics data analysis and experiments. However, pipelines often lack standardization and portability, hindering widespread adoption and collaboration. In addition, owing to pipeline component updates and the need for specialized compute environments, reproducibility is a concern. As datasets grow larger and become more complex, scaling pipelines efficiently also becomes even more challenging, often requiring the transition to more powerful compute infrastructure or cloud-based environments. Pipelines must be able to handle these increasing demands while ensuring efficient data processing and analysis, which can be resource-intensive. This is why pipeline modularity and interoperability are key, akin to microservices in software.
💡Consideration: Scalable workflows
✔ Implement automated workflows with workflow management systems (e.g., Nextflow).
✔ Use containerized workflows and source code management systems.
✔ Test and verify pipelines with code-checking tools, such as Seqera AI.
✔ Leverage cloud-based and HPC resources to dynamically scale and optimize compute.
4. Analytics and Compliance
Bioinformatics is not just about running a single pipeline. Rather, it involves the establishment of reporting and provenance tracking to ensure reproducible pipeline executions. Data across many fields suggests that reproducibility is lower than desirable, with one study estimating that 85% of biomedical research efforts are wasted. As datasets become more intricate and pipelines grow increasingly complex, ensuring transparency and traceability of every step becomes an even greater challenge. The volume and diversity of data, combined with the evolving nature of bioinformatics workflows, make it essential to implement robust and automated monitoring systems that track provenance, ensure compliance, and provide visibility into resource usage. We can leverage observability and audit logging concepts and tools from DevOps (e.g., Grafana, AWS Cloud Watch).
💡Consideration: Establish Monitoring Systems
✔ Audit access and logging for traceability and compliance.
✔ Implement automated monitoring with notifications.
✔ Manage infrastructure with cost controls and resource tagging.
5. Interactive Analysis
Data analysis extends beyond running pipelines, with downstream analysis and human interpretation often required for reporting and extracting meaningful scientific insights. Interactive analysis tools like dashboards, graphs, and visualizations enable researchers to further explore results and refine hypotheses. However, performing tertiary analysis is considered one of the most challenging steps in the entire bioinformatics process. In order to perform further analysis on pipeline outputs, researchers often waste time transferring data between cloud and local storage. Additionally, researchers often need to navigate the complex landscape of multiple libraries, tools, and programming languages. This fragmented process increases the potential for errors, ultimately slowing down research progress. Moving towards real-time, cloud-native analysis, similar to stream processing in data engineering.
💡Consideration: Interactive Analysis Environments
✔ Run interactive analysis in the same location as your data (Seqera Studios, Posit, GitHub)
✔ Leverage reproducible, containerized interactive environments (e.g., Jupyter and R IDE notebooks, VS Code IDEs, Xpra remote desktops).
✔ Centralize data, pipelines, and interactive analysis into a unified platform (e.g. Seqera).
Moving Towards a Unified, Scalable Data Lifecycle
By adopting unified, scalable platforms and interactive tools, researchers can streamline workflows, improve collaboration, and ensure reproducibility. Addressing these challenges will accelerate research, enabling more impactful discoveries and driving innovation across scientific fields.
Want to get more insights? Subscribe to our monthly newsletter
