BigOmics Meets Studios: End-to-end Exploration of RNA-Seq and Proteomics Data in Seqera Platform
Omics data is growing at an unprecedented pace. Databases like GenBank and SRA already hold over 100+ petabytes of data and are expected to surpass 2.5 exabytes (2.5 billion gigabytes) this year. The bottleneck is no longer generating data, but analyzing it. How can scientists ensure reproducible data analysis at scale, across diverse datasets, analysis tools, and rapidly evolving computational landscapes?
BigOmics Analytics have partnered with Seqera to bring Omics Playground (BigOmics’ interactive RNA-seq and proteomics workbench) into Studios (the interactive analysis feature of Seqera Platform). The result is a seamless, end-to-end path from raw reads to interactive plots, all deployed on the same infrastructure as your Nextflow pipelines. In this post, we’ll explore how this integration is enabling a scalable and secure solution for omics, featuring a drug resistance cancer use case.
Request a Studios DemoOmics Playground: Interactive Multiomics Exploration
Omics Playground is a data discovery and exploration platform for single and multiomics that enables dry-lab bioinformaticians to interactively share analysis results with wet-lab bench scientists and biologists. It simplifies the analysis, interpretation, and presentation of complex biological data, enabling bioinformaticians to offload repetitive tasks and focus on higher-level insights, whilst giving biologists transparency and a deeper understanding of the data.
With Omics Playground, bioinformaticians can easily package QC metrics, differential expression, pathway enrichment, and biomarker discovery into interactive, no-code dashboards. Omics Playground also includes curated gene-set libraries, point-and-click visualizations (PCA, volcano plots, GSEA, drug-cMAP), and shareable workbooks—so insights travel as fast as the questions behind them.
Key Benefits of Omics Playground
- →Centralized and searchable data management: Seamlessly manage and compare multiple datasets, accelerating discovery over time.
- →Standardized, reproducible results: Apply consistent statistical frameworks across studies to ensure robust and reliable outcomes.
- →Efficient and scalable: Analyze large-scale datasets up to 10x faster, dramatically reducing time from raw data to insight.
- →Publication-ready visuals: Export high-quality, customizable figures perfect for presentations, publications, and team collaboration.
💡Interested in Omics Playground? See it in action: Request a Guided Tour Now!
Studios: Reproducible, On-Demand Analysis Environments
Studios, a feature of Seqera Platform, streamlines the entire data lifecycle, closing the loop between data, Nextflow pipelines, and downstream analysis. It provides real-time access to reproducible, containerized environments—empowering users to explore results instantly using familiar bioinformatics tools. With Studios, you can launch containerized IDEs such as Jupyter, R-IDE, VS Code, or a full Linux desktop—co-located with your cloud data. Credentials, object-store throughput optimization, session lifespans, and session checkpointing are handled automatically. If you need a custom application, just ship your own Docker image, and Studios runs it seamlessly.
Key Benefits of Studios
- →One-click data co-location: Mount data in object storage directly inside the Studio workspace; no rsyncs or presigned URLs. Interact with cloud data as if it were local.
- →Reproducible, version-controlled containers: Every Studio is a tagged container image, ensuring consistent, reproducible setups that can be reopened or shared anytime.
- →Customizable and extensible: Use pre-built templates, augment an existing template, or bring your own Docker container template image. Or even mount your own application stack via EFS. Studios adapt to your specific tools, libraries, and analysis needs.
- →Real-time collaboration: Invite colleagues via a simple URL; multiple users can work in the same notebook or dashboard simultaneously for live cross-validation of findings.
- →Optionally limit lifespans for cost: Sessions can run indefinitely, or have a defined expiration time. Sessions can be extended ad-hoc as needed.
- →Private or shared access: Administrators can choose whether sessions are collaborative or sandboxed for a single user.
💡Want to learn more about Studios and Seqera Platform? Request a demo now
Why Integrate Omics Playground into Studios?
Omics analysis demands an end-to-end solution that combines advanced tools and scalable, secure infrastructure. Integrating Omics Playground within Studios addresses this need by enabling collaborative, interactive RNA-Seq and proteomics analysis right where your data lives. Scientists can seamlessly transition from running Nextflow pipelines to downstream exploration with Omics Playground, all within a single unified platform.
Traditional workflows require downloading large result files, setting up local GUIs, and manually syncing outputs. With Omics Playground running inside Studios, those hand-offs and time sinks are eliminated:
| Challenge | Studios and Omics Playground Integration |
|---|---|
| Moving multi-GB count matrices off-cloud | Data stays in object storage. Seqera’s Fusion filesystem streams it into the container on demand. |
| Reproducing a colleague’s interactive session | Launch the same container tag and snapshot to regenerate interactive plots automatically. |
| Scaling CPU/GPU needs mid-analysis | Dynamically resize the Studio’s compute profile without needing to restart the app. |
| Ensuring security and compliance | Interactive sessions inherit IAM policies, network controls, and audit logging from the compute layer. |
💡Hint: Fusion is an optimized file system designed for cloud-native pipelines enabling analysis of data-in-place at any scale. Fusion fully optimizes how Nextflow uses data – enabling you to simultaneously increase performance, lower costs, simplify infrastructure, and improve reliability.
Real-world Example: Revisiting Glucocorticoid Resistance in Multiple Myeloma
💡Prerequisites: Studios is now generally available in Seqera Platform v25.1 and later. To get started, ensure you’re running the latest version of the platform.
Drug resistance is a major challenge in Multiple Myeloma (MM), a hematological cancer that frequently develops resistance to anticancer drugs. In this example, we demonstrate how to use the Omics Playground integration with Studios to perform end-to-end analysis on PRJNA681920, a public dataset containing mRNA profiles of glucocorticoid-sensitive (MM1S) and glucocorticoid-resistant (MM1R) B lymphoblast cell lines after treatment with withaferin or DMSO (UT, untreated control).
1. Gather the data
For this particular dataset, the processed counts files are already available on GEO, but to emulate a full experiment from raw fastq.gz to biological insight, you might pull raw reads from SRA and align them yourself using Nextflow. The nf-core/fetchngs pipeline will pull data from SRA and save the raw fastq files in your own infrastructure ready for mapping and normalization.
2. Run your Nextflow pipeline
To analyze gene expression changes related to glucocorticoid resistance, we can start by processing the raw sequencing data. The nf-core/rnaseq pipeline can perform alignment, quantification, and DESeq2-based normalization. The workflow provides you clean, reproducible gene expression counts for downstream analysis.
3. Launch a Studio session
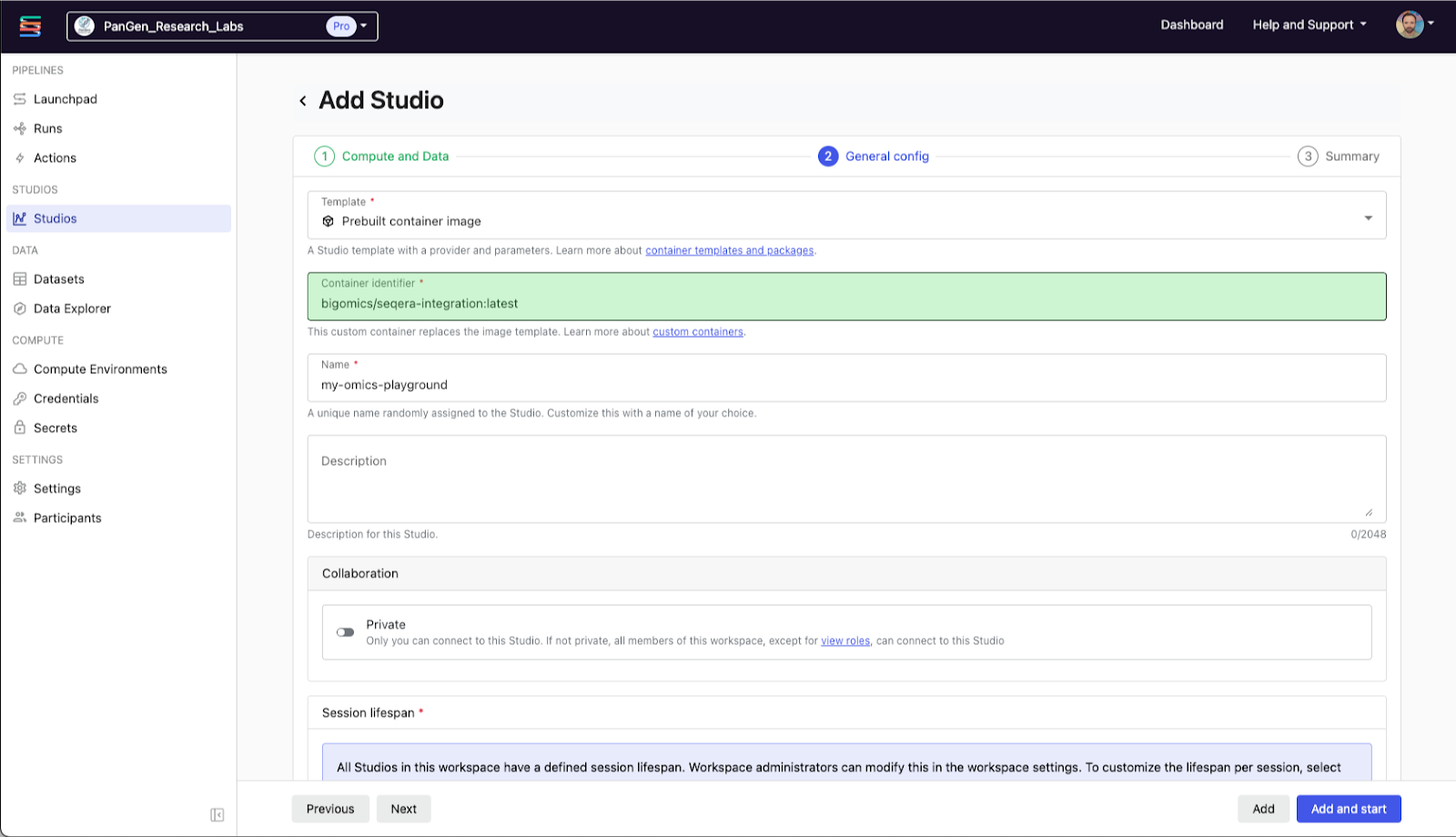
Once the pipeline completes, the counts files will be written to your bucket. When creating the Studio, you can select the nf-core/rnaseq workflow’s output directory (or a parent directory) to be mounted inside your session. Studios provide an interactive, containerized workspace co-located with your data so you can start exploring results immediately, without downloading files or switching environments. When choosing the Studio template, choose “Prebuilt container image” and enter bigomics/seqera-integration:latest as shown below:
4. Interactively explore results in Omics Playground
Click the blue “Add and start” button to launch Omics Playground directly within your Seqera Platform workspace. The Omics Playground application will be hosted in your own infrastructure. The Fusion filesystem provides input data such as counts matrices and sample sheets to the Omics Playground so that they can be loaded into an interactive dashboard with PCA plots, heatmaps, GSEA modules, ready for immediate exploration without creating a second copy of the data in a new location.
5. Share and collaborate in real time
Generate a shareable link to your Studio session so collaborators can join the same live environment - no need to rerun pipelines or set up tools. They can filter results, explore drug-cMAP correlations, bookmark DE tables, and contribute insights in a shared, reproducible workspace.
Interested in learning more about Studios? Request a Demo or watch the full webinar on demand

