Accelerating Analytics with Easy Genomics - Wisconsin State Laboratory
By: Venkata Kampana, Senior Solutions Architect, AWS and Dr. Dawn Heisey-Grove, Public Health Analytics Leader, AWS.
Genomics is the study of the structure and function of an organism’s genome and uses sequence information to better understand the organism and the effects of evolutionary changes that occur over time. Advances in sequencing and computational technologies are enabling public health agencies (PHAs) to apply genomics towards a better understanding of disease and disease prevention. However, public health agencies (PHAs) may be challenged by limited access to bioinformaticians trained to use current tools.
Covid highlights the need for a more accessible open source solution for public health agencies
Among the many things that public health agencies (PHAs) learned during the pandemic is that timely and complete access to viral genomic sequencing is critical to genetic surveillance and informing public policy. PHAs need to know when a new SARS-CoV-2 variant is increasing in prevalence. When paired with clinical and epidemiologic data, this information can be used to understand which variants might cause more disease in a community. It can also provide an early warning of new variants that may cause more severe illness or death.
On-demand access and elasticity of the Amazon Web Services (AWS) cloud is well matched to meet the increasing demands for genomic data storage and compute in public health settings. However, PHAs need the capability to rapidly scale capacity by enabling existing lab technicians without a background in informatics or cloud operations to run genomics pipelines in the cloud. To accomplish this, PHAs need an easy to use solution tailored to their specific set of requirements.
Democratizing public health access to pipelines and data
The Wisconsin State Laboratory of Hygiene (WSLH) wanted the ability to scale genomic sequencing analytics capacity by making bioinformatics workflows more accessible to laboratories with limited bioinformatics capacity. To create an open-source solution that could be freely shared with other public health agencies, WSLH decided to work with Two Bulls (Part of DEPT®). Two Bulls, a member of the AWS Partner Network (APN), is skilled at developing open source, intuitive websites and user interfaces and has considerable experience developing solutions for government services, especially health-related services.
Developed through that collaboration between WSLH and Two Bulls, the Easy Genomics solution was designed to provide a simple interface for uploading genomic sequencing data to the cloud and running (pre-configured) genomics pipelines via NextFlow Tower’s APIs.
To avoid reinventing the wheel, WSLH needed the capability to leverage independently developed pipelines widely used in public health. Many of these pipelines are available as part of the nf-core project — a community effort to collect a curated set of analysis pipelines built using Nextflow such as ampliseq, mag, viralrecon, and rnaseq. With Easy Genomics, non-technical public health staff can upload genomic sequences and run the right pipelines with a few clicks.
What is Easy Genomics?
Easy Genomics is designed for organizations that need a simple browser interface for lab technicians or other staff to upload raw genomic sequences from processed samples to the AWS cloud for processing using Seqera Lab’s NextFlow Tower solution. Easy Genomics presents an easy landing page for individuals to:
- Securely login;
- Upload data from single or multiple samples;
- Select the appropriate analytic workflow;
- Monitor sample progress; and
- Receive the results via the browser or email.
To make the experience seamless for its users, Easy Genomics includes functionality that allows an Administrator to:
- Add, modify, or delete users;
- Add, modify, or delete organizations; and
- Manage users’ pipeline access.
Two Bulls plans to release Easy Genomics as an open-source offering with Cloud Formation and Terraform templates to deploy the solution in an AWS environment. Labs and PHAs can optionally customize the web interface (including their own text and logos) using an administration tool provided as part of the distribution.
Easy Genomics uses Nextflow and NextFlow Tower APIs for access to core services such as managing compute environments, users, groups, pipelines, and providing end-to-end workflow processing.
Nextflow Tower
Nextflow Tower is a web-based command post that enables users to launch, manage, monitor and collaborate on Nextflow data analysis pipelines on-premises or in the cloud. Tower enables organizations to:
- Launch and manage scalable data analysis pipelines from a secure command post;
- Enable pipeline portability across cloud and on-prem HPC environments;
- Facilitate collaboration between local and distributed teams;
- Help meet regulatory requirements with reliable, predictable, reproducible, and auditable pipeline execution.
While Tower provides its own Web UI tailored to administrators and bioinformaticians, it also exposes a public API with necessary endpoints to manage Nextflow workflows, compute environments, credentials, users, and other Tower objects. This API enables services in Tower to be easily accessible to external applications such as Easy Genomics.
A scalable managed architecture
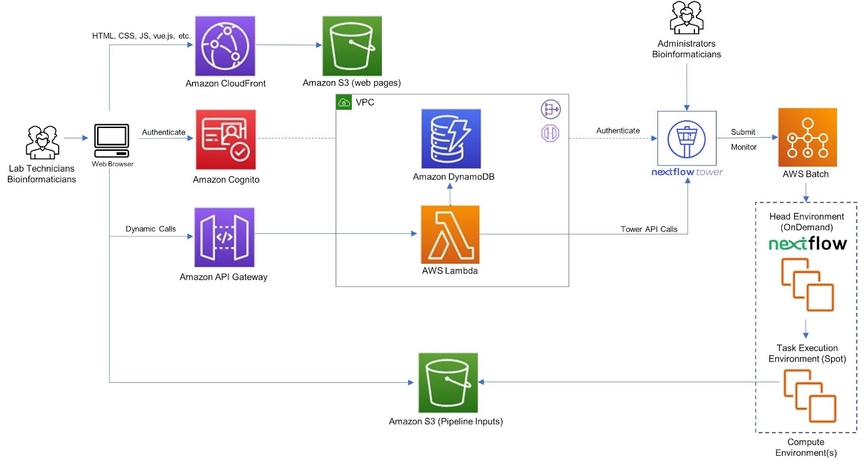
This reference architecture below provides an overview of Easy Genomics and how it integrates with Nextflow Tower. The guidance provided here is generic and must be adapted to meet specific customer requirements and desired business outcomes.
Easy Genomics uses AWS serverless services, which provide automatic scaling and built-in high availability. Serverless services used in Easy Genomics include AWS Lambda, Amazon API Gateway, Amazon DynamoDB, Amazon Cognito, Amazon S3, Amazon Cloudfront, and AWS Batch. Easy Genomics stores static web resources—including HTML, CSS, JavaScript, and image files—in Amazon S3, and serves them to the individual’s browser via CloudFront content delivery network (CDN) service. This creates a secure, cost-effective front end that scales as demand and workload changes. Easy Genomics also displays the analytic results in this browser front end and can send them via email.
The front-end application sends and receives data from a back-end micro-service built using AWS Lambda and the Amazon API Gateway. AWS Lambda functions invoke the NextFlow Tower API to execute the pipelines. The Tower API requires an authentication token to be specified in each API request using the Bearer HTTP header. Administrators can generate that token from the Nextflow Tower. Information on how to generate that token is provided in the Nextflow Tower API Overview documentation.
Laboratorians access Easy Genomics using the web browser, upload the sequencing data, select the workflow and submit it for processing. Behind the scenes, Easy Genomics interfaces with Nextflow Tower APIs for processing the workflows and upload the results back to an Amazon S3 Bucket.
In the reference architecture above, calls are made to the Nextflow Tower service at https://tower.nf. Commercial Nextflow Tower customers can elect to use hosted Tower service, or they may optionally deploy Tower within their own AWS environments. Tower components are packaged as Docker container images which are hosted and security validated by the Amazon ECR service. Tower can be deployed in AWS as a set of containers or optionally as a service managed by Amazon EKS.
Nextflow Tower provides a built-in integration with AWS Batch and other compute environments. AWS users simply store their AWS Batch and storage credentials securely in Tower, and Tower automatically creates and manages compute resources in AWS to execute multiple pipelines concurrently.
Read Nextflow and AWS Batch – Inside the Integration
Built with data security in mind
Two Bulls ensured that data security is at the forefront in Easy Genomics, which uses Amazon Cognito for user management and authentication to secure the REST API in Amazon API Gateway. User accounts are stored in Amazon Cognito, with site Administrators managing user accounts and delegating access to perform various workflow activities (create, update, delete, run workflows) through the web interface. Easy Genomics manages authentication to Nextflow Tower via an API key.
Easy Genomics stores the metadata about the labs and pipeline runs in Amazon DynamoDB. DynamoDB is a fast and flexible non-relational database service for any scale. It provides a highly durable storage infrastructure designed for mission-critical and primary data storage. Data are redundantly stored on multiple devices across multiple facilities in an Amazon DynamoDB region. Similarly, Amazon Simple Queue Service (Amazon SQS) offers a secure, durable, and available hosted queue to integrate and decouple distributed software systems and components. Easy Genomics uses SQS to support redundancy if the Nextflow Tower API is unavailable.
To store lab pipeline input and output artifacts, Easy Genomics allows administrators to apply all security best practices to data storage of the uploaded samples and stored results (e.g., storage in new S3 buckets, data encryption, versioning).
Run DynamoDB and Lambda functions in private subnets within a virtual private cloud (VPC). Lambda functions, which are deployed in a private subnet, access the Nextflow Tower Public APIs by using a NAT Gateway. Use VPC endpoints for communication between Lambda and Amazon SQS queue, AWS Batch (where workflow jobs will be executed), and Amazon S3.
Enabling scalable data analysis
Easy Genomics solves a critical gap in public health laboratories’ genomics sequencing ability to scale in times of crisis. Public health laboratories require a genomic sequencing solution that is easy to use, manage, and scale. With the combination of Easy Genomics and Nextflow Tower, public health agencies can:
- Easily manage lab sequencing datasets and launch pipelines with minimal training or expertise;
- Enable secure collaboration to boost productivity;
- Rapidly scale analysis capability to meet unexpected demands using AWS cloud platform;
- Minimize infrastructure costs by only paying for infrastructure when it is required.
Two Bulls founder Evan Davey says, “All through the COVID-19 pandemic, Two Bulls supported the response of the Government of Victoria with our full tech stack tech expertise. Democratizing access to genomic sequencing is the next logical step, and we’re thrilled to see this solution come together.”
Rob Lalonde, chief commercial officer at Seqera says, “We’re delighted to see Nextflow Tower being used in new and creative ways to solve real-world problems. Easy Genomics presents an exciting new opportunity to make pipelines and data more accessible to end users while realizing the benefits of scalable, cost-efficient pipeline execution on AWS infrastructure.”
Recommendations when using Easy Genomics
- Encrypt data at rest using customer managed keys for least-privileged access controls and AWS Key Management Service (KMS);
- Configure compute environments in Tower to use the AWS Batch integration. Tower Forge automatically provisions Amazon Elastic Compute Cloud (Amazon EC2) in AWS Batch so that organizations only pay for the infrastructure they use;
- Through the Tower interface, administrators can optionally configure a compute environment that uses a high-performance Amazon FSx for Lustre file systems for data-intensive pipelines;
- Keep costs low by using Amazon EC2 Spot Instances configurable in Nextflow Tower, which can reduce your cost by as much as 90% compared to on-demand compute prices. Nextflow Tower has the ability to automatically relaunch failed tasks, you can benefit from Spot Instances’ lower costs without concern for losing your tasks and the data associated with it;
- Use Amazon S3 Glacier for long-term storage of infrequently used data. S3 Glacier provides Long-term, secure, durable storage classes for data archiving at the lowest cost and milliseconds access. Customers can use Glacier to store both raw sequencing datasets and outputs generated by Nextflow pipelines;
- Organizations using Easy Genomics and Nextflow Tower in production might consider hosting Tower in their own AWS environment to reduce latency and simplify management.
Next steps
Interested in learning more about the resources we mentioned above?
Learn more about Nextflow Tower go to: https://seqera.io/tower/
To discuss the commercial licensing of Nextflow Tower go to: https://seqera.io/demo/
To learn more about other open source solutions developed by Two Bulls go to: https://www.twobulls.com/
A variety of high-quality curated Nextflow pipelines that work with Easy Genomics and Nextflow Tower are available at https://nf-co.re/.
Learn more about AWS solutions for public health and genomics at: https://aws.amazon.com/health/genomics/