Streamlining containers lifecycle
In the bioinformatics landscape, containerized workflows have become crucial for ensuring reproducibility in data analysis. By encapsulating applications and their dependencies into portable, self-contained packages, containers enable seamless distribution across diverse computing environments. However, this innovation comes with its own set of challenges such as maintaining and validating collections of images, operating private registries and limited tool access.
Seqera’s Wave tackles these challenges by offering a suite of features designed to simplify the configuration, provisioning and management of software containers for data pipelines at scale. In this blog, we will explore common pitfalls of managing containerized workflows, examine how Wave overcomes these obstacles, and discover how Seqera Containers further enhances the Wave user experience.
Read the Whitepaper Now!
Handling containerized workflows at scale is not easy
Software containers have been heavily adopted as a solution to streamline both the configuration and deployment of dependencies in complex data pipelines. However, maintaining containers at scale is not without its difficulties. Building, storing and distributing container images is an error-prone and tedious task that increases the cognitive load on software engineers, ultimately diminishing their productivity. Community-maintained container collections, such as BioContainers, have emerged to mitigate some of these challenges. However, still, several problems remain:
- →Publicly Accessible Container Images: Issues with stability can compromise reliability. Typically unsuitable for non-academic organizations due to security and compliance concerns.
- →Limited Tool Access: Access is restricted to only to specific tools or collections (e.g. BioConda). Organizations often need the flexibility to assemble and deploy custom containers.
- →API Rate Limits: Public registries often impose low API rate limits and afford low-rate or low-quality SLAs, making them unsuitable for production workloads.
- →Egress Costs: Use of private registries can incur outbound data transfer costs, particularly when deploying pipelines at scale across multiple regions or cloud providers.
Seqera’s Wave solves these problems by simplifying the management of containerized bioinformatics workflows by provisioning containers on-demand during pipeline execution. This approach ensures the delivery of container images that are defined precisely depending on requirements of each pipeline task in terms of dependencies and platform architecture. The process is completely transparent and fully automated, eliminating the need to manually create, upload and maintain the numerous container images required for pipeline execution.
By integrating containers as dynamic pipeline components rather than standalone artifacts, Wave streamlines development, enhances reliability, and reduces maintenance overhead. This makes it easier for developers and operations teams to build, deploy, and manage containers efficiently and securely.
How does Wave work?
Wave transforms containers and pipeline management by allowing bioinformaticians to specify container requirements directly within their pipeline definitions. Instead of referencing manually created container images in Nextflow’s container directive, developers can either include a Dockerfile in the directory where the process' module is defined or just instruct Wave to use the Conda package associated with the process definition. By using this information, Wave provisions a container on-demand either using an existing container image in the target registry matching the specified requirement or building an new one on-the-fly to fulfill a new request, and returns the container URI pointing to the Wave container for process execution. The built container is then pushed to a destination registry and returned to the pipeline for execution, ensuring seamless integration and optimization across diverse computational architectures.
Wave can also direct containers into a registry specified in the nextflow.config file, along with other pipeline settings. This means containers can be served from cloud registries closer to where pipelines are executed, delivering better performance and reducing network traffic. Moreover, Wave operates independently, serving as a versatile tool for bioinformaticians across various platforms and workflows. By employing multi-level caching, Wave ensures that containers are built only once or when the Dockerfile changes, enhancing efficiency and streamlining the management of bioinformatics workflows.
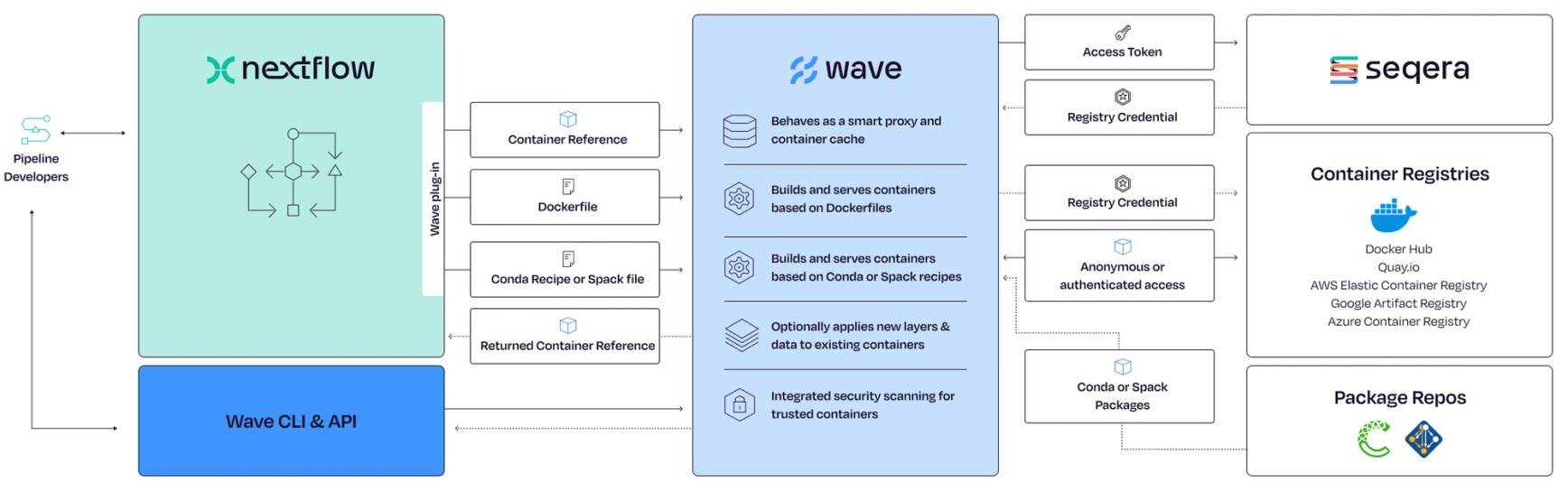
Figure 1. Wave —a smart container provisioning and augmentation service for Nextflow.
Key features of Wave
✔ Access private container repositories: Seamlessly integrate Nextflow pipelines with Seqera Platform to grant access to private container repositories.
✔ On-demand container provisioning: Automatically provision containers (via Dockerfile or Conda packages) based on dependencies in your Nextflow pipeline, enhancing efficiency, reducing errors, and eliminating the need for separate container builds and maintenance.
✔ Enhanced security: Each new container provisioned by Wave undergoes a security scan to identify potential vulnerabilities.
✔ Create multi-tool and multi-package containers: Easily build and manage containers with diverse tools and packages, streamlining complex workflows with multiple dependencies.
✔ Provision multi-format and multi-platform containers: Automatically provision containers for Docker or Singularity based on your Nextflow pipeline configuration and platform, including ARM64 containers for AWS Graviton if a compatible Dockerfile or Conda package is provided.
✔ Mirror Public and Private Repositories: Mirror the containers needed by your pipelines in a registry co-located with where pipeline execution is carried out, allowing optimized data transfer costs and accelerated execution of pipeline tasks.
Download the Whitepaper to explore features in more detail
Seqera Containers for publicly accessible container images
With the newly launched Seqera Containers, the Wave experience is elevated even further. Now, instead of browsing existing container images as with a traditional container registry, users can just specify which tools they require through an intuitive and user-friendly web interface. This will find an existing container image for the required tool(s) or build a container on-the-fly using the Wave service. Currently it supports any software package provided by the Bioconda, Conda forge and Pypi Conda channels. Container can be built both for Docker and Singularity image format and linux/amd64 and linux/amd64 CPU architecture.
Additionally, Seqera Containers are stored permanently and publicly accessible via the registry host community.wave.seqera.io. This ensures that any future requests for the same package will return the exact container image, guaranteeing reproducibility across runs. Seqera Containers project was developed in collaboration with Amazon Web Service, which is sponsoring the container hosting infrastructure.
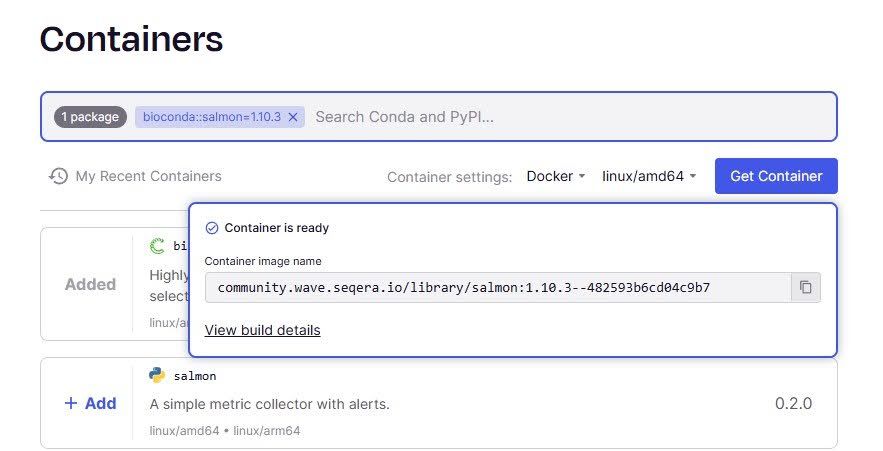
Figure 2. Snapshot of Seqera Containers, demonstrating how you can create containers with the tools you want, on the fly.
Discover the benefits of Wave
Wave offers a transformative solution to the complexities of managing containerized bioinformatics workflows. By integrating containers directly into pipelines and prioritizing flexibility and efficiency, Wave streamlines development, enhances security, and optimizes performance across diverse computing environments. Deep dive into how Wave can revolutionize your workflow management by downloading our whitepaper today.
Download the Wave Whitepaper
