This post has been written by our valued community members.
Removing bioinformatics bottlenecks in pathogen surveillance is crucial for public health, enabling early detection and response to emerging infectious threats. VIRUS-MVP leverages Nextflow and nf-core to provide a modular, automated, and scalable framework for genomic surveillance of priority viruses. Designed initially for SARS-CoV-2, the workflow was later adapted or is in development for other priority viruses, including Mpox, respiratory syncytial virus (RSV), and influenza.
What is VIRUS-MVP?
VIRUS-MVP is an open-source, pathogen-agnostic bioinformatics workflow that helps scientists detect and track viral mutations. By integrating sequencing data with curated biological annotations, it enables public health labs and researchers to understand how viruses are changing over time and what those changes might mean for disease spread, immunity, or treatment effectiveness.
The power of Nextflow and nf-core
One of the key strengths of VIRUS-MVP is its plug-and-play approach powered by Nextflow and nf-core. These technologies enable pipeline reproducibility, scalability, and interoperability, making it easier to integrate new pathogens into the workflow. Additionally, the workflow is highly adaptable to different data types, such as clinical sequencing data and wastewater samples, ensuring comprehensive surveillance. The Nextflow architecture also allows users to switch between different bioinformatics tools based on data requirements—for example, selecting appropriate variant callers depending on sequencing depth and quality.
Nextflow’s Channel factories and Operators are crucial in processing diverse data types. These features facilitate the dynamic grouping of data based on different criteria, such as time-series trends or geographical clustering, enabling researchers to analyze pathogen spread and evolution more precisely.
Furthermore, containerization (e.g., Docker and Singularity) enhances workflow portability, enabling execution across various computing environments, from local workstations to cloud-based infrastructure. This portability allows provincial and federal public health laboratories to deploy the framework locally, ensuring a bringing-tools-to-data approach that enhances security and data sovereignty. The use of Wave containers from Nextflow further accelerates development by enabling rapid testing and integration of new bioinformatics modules without modifying the entire workflow.
Workflow overview
VIRUS-MVP follows a structured multi-step approach:
- Preprocessing: Standardizes input files and prepares them for downstream analyses.
- Quality Control: Assesses sequencing data quality.
- Mapping: Aligns reads to reference genomes using industry-standard tools.
- Variant Calling: Identifies mutations in viral genomes.
- Annotation: Provides functional insights into detected variants.
- Surveillance Reporting: Generates structured outputs for epidemiological insights.
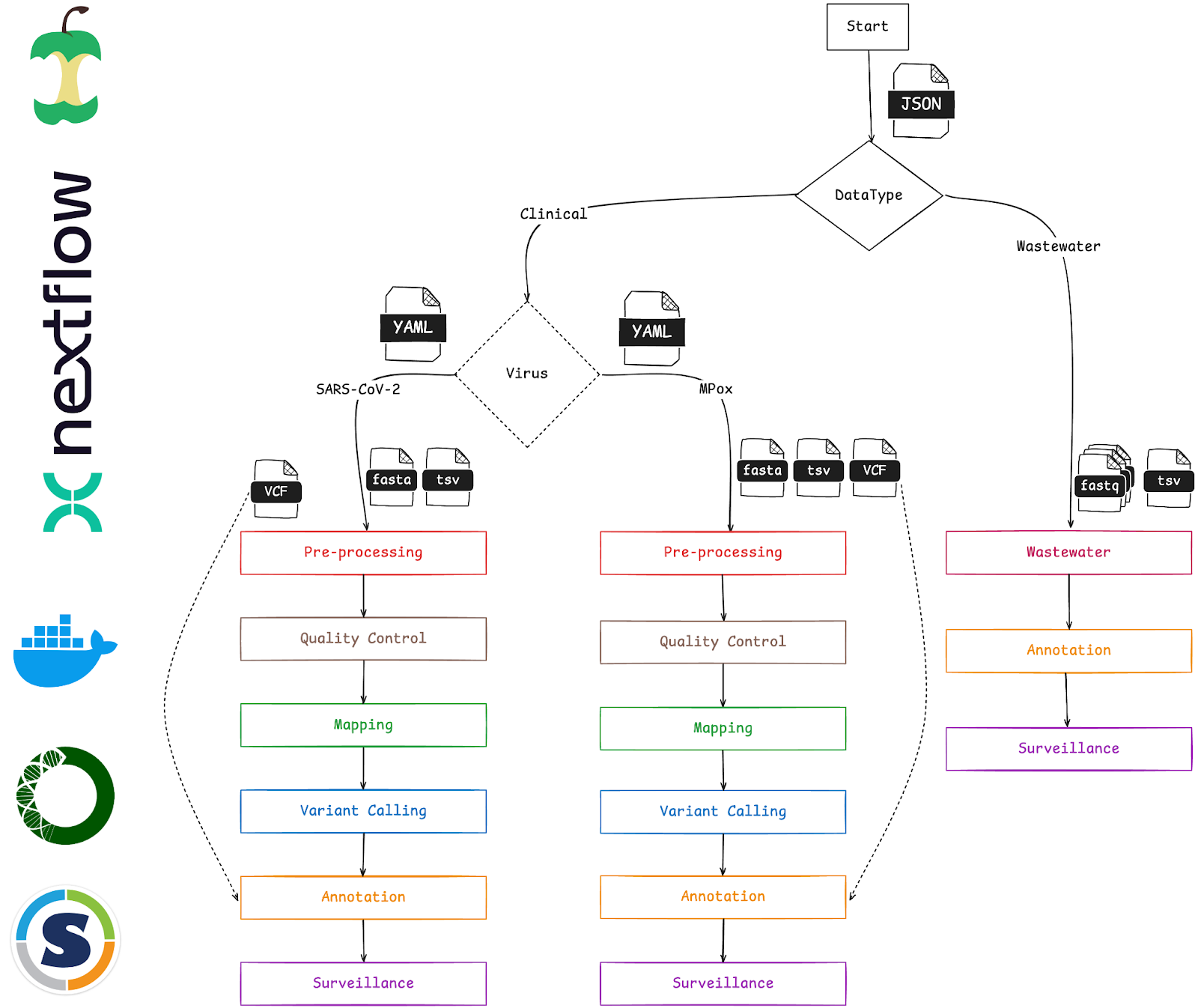
Figure 1. Overview of the modular and scalable VIRUS-MVP workflow for pathogen surveillance, illustrating the seamless integration of key components from data input to final reporting.
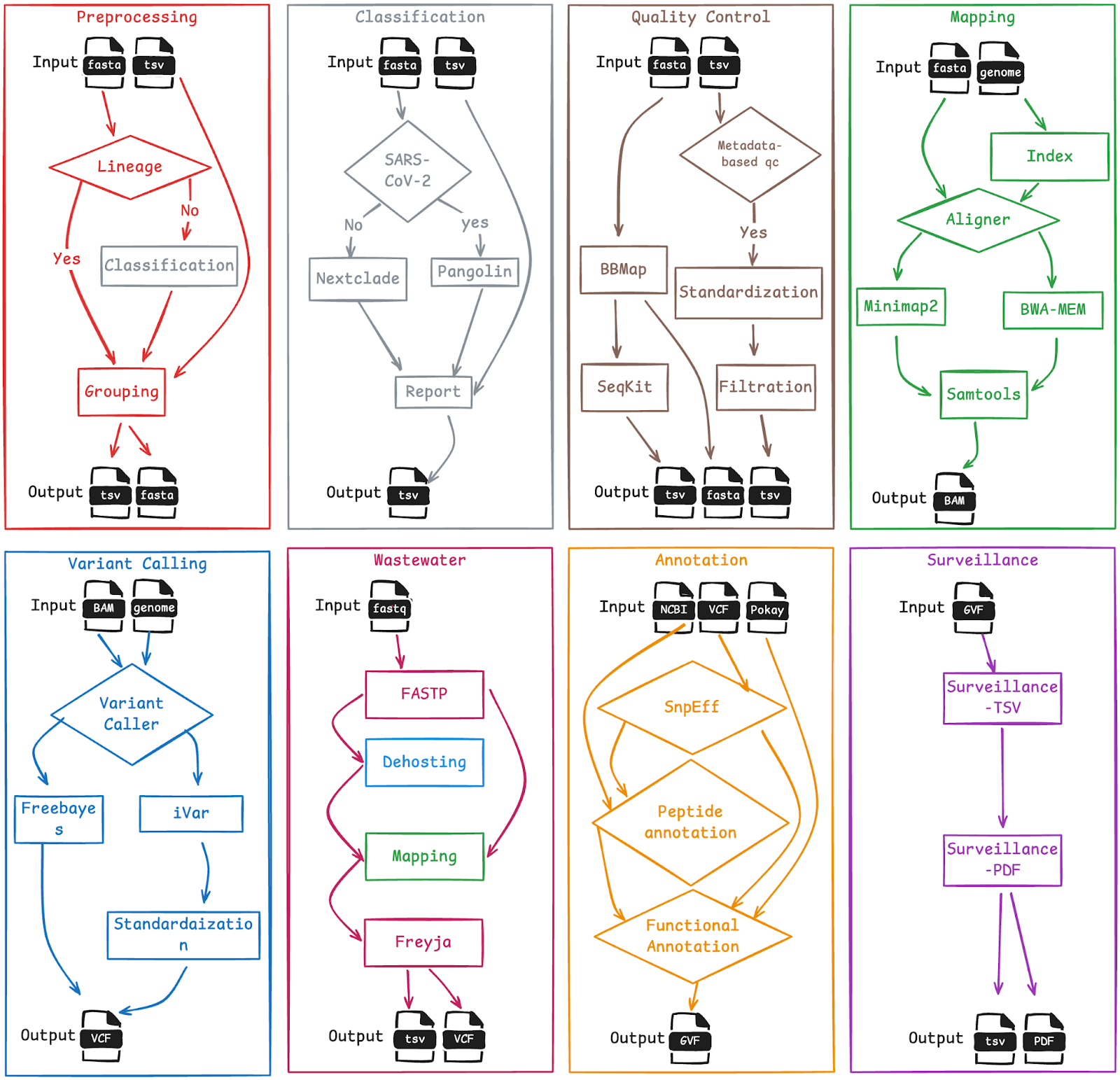
Figure 2. Detailed view of the individual sub-workflows that constitute the overall VIRUS-MVP workflow, highlighting the modular structure and flexible architecture enabling customization and reuse across diverse surveillance contexts.
Real-World Applications of VIRUS-MVP
VIRUS-MVP is actively used in real-world genomic surveillance efforts. In Canada, it has been deployed for SARS-CoV-2 surveillance, leveraging sequencing data available through the Canadian VirusSeq Data Portal. This implementation enables continuous monitoring of viral mutations and variant emergence across the country.
Additionally, a dedicated instance for mpox surveillance has been developed utilizing data from Pathoplexus. This adaptation demonstrates the flexibility of VIRUS-MVP in accommodating different pathogens, providing a robust framework for genomic epidemiology and public health decision-making.
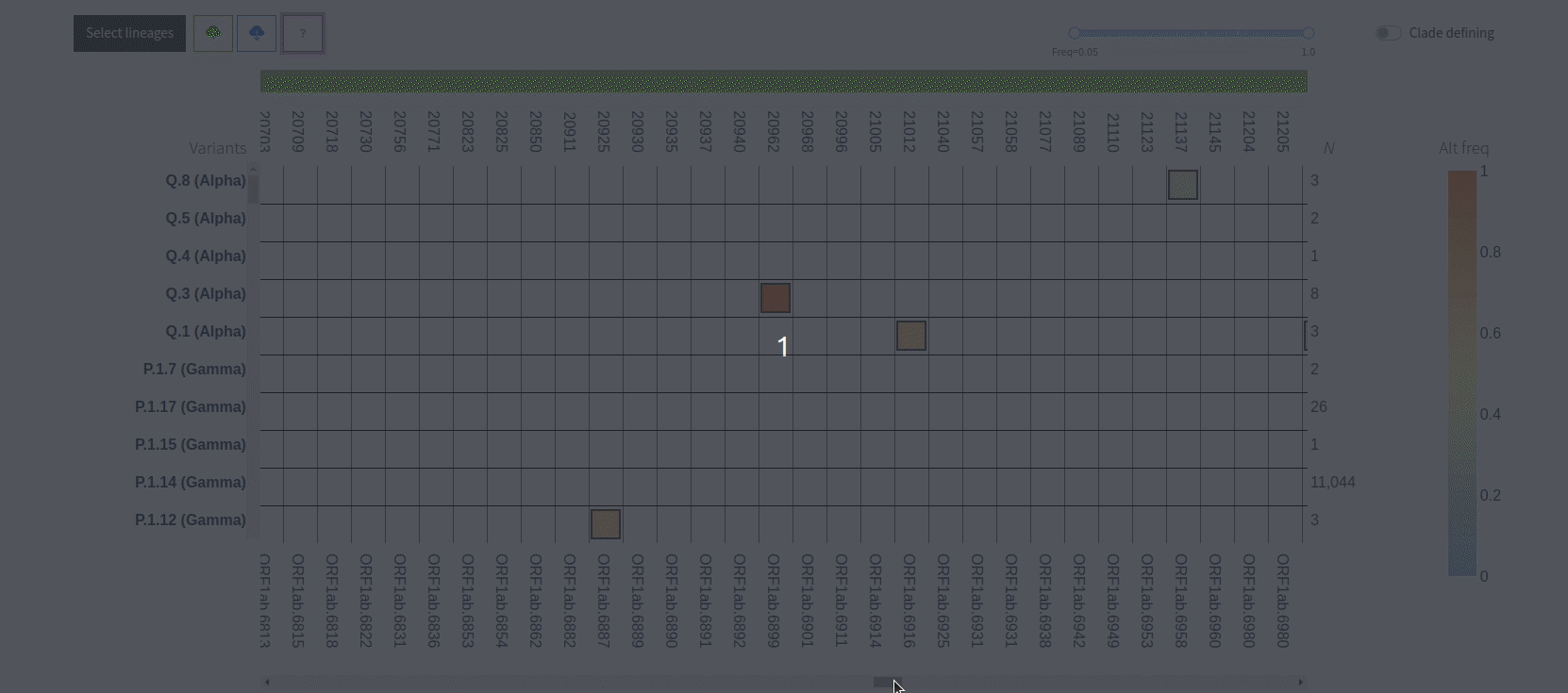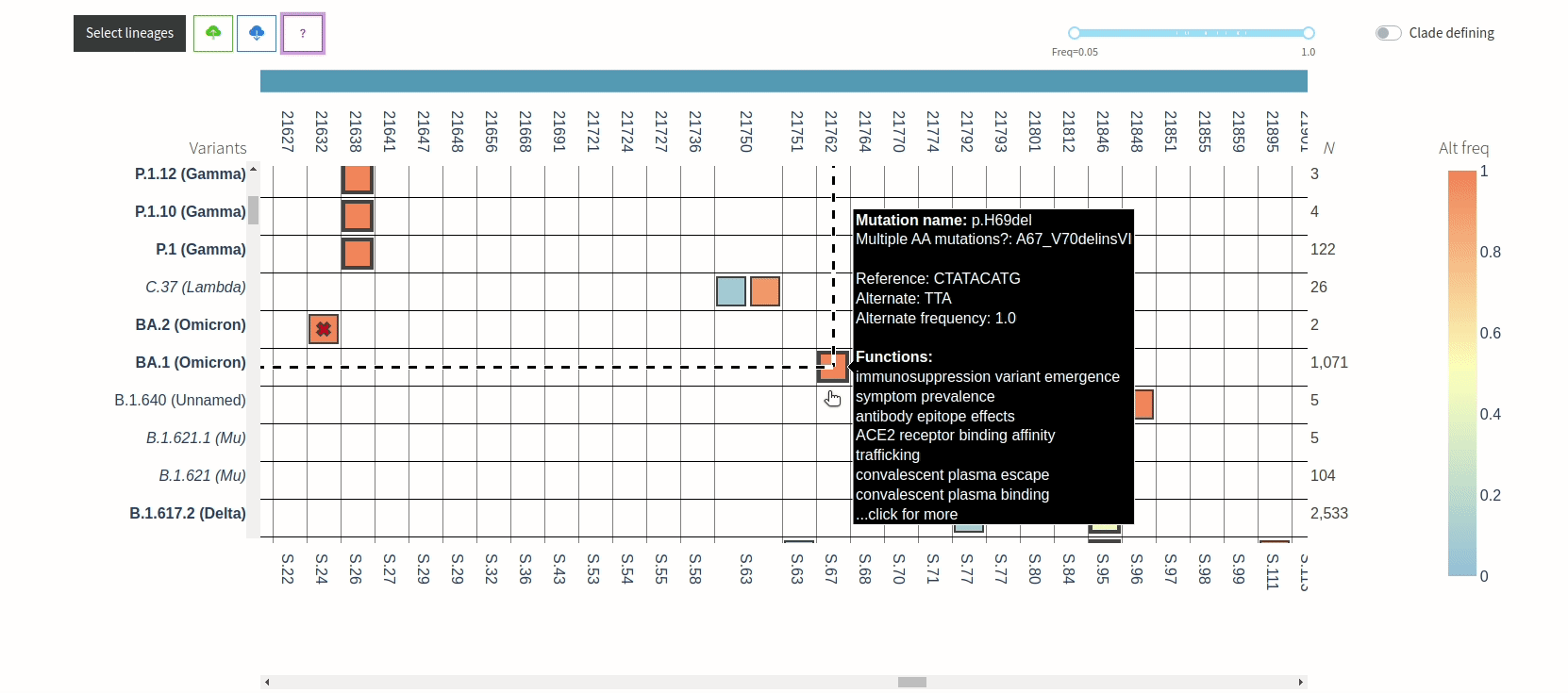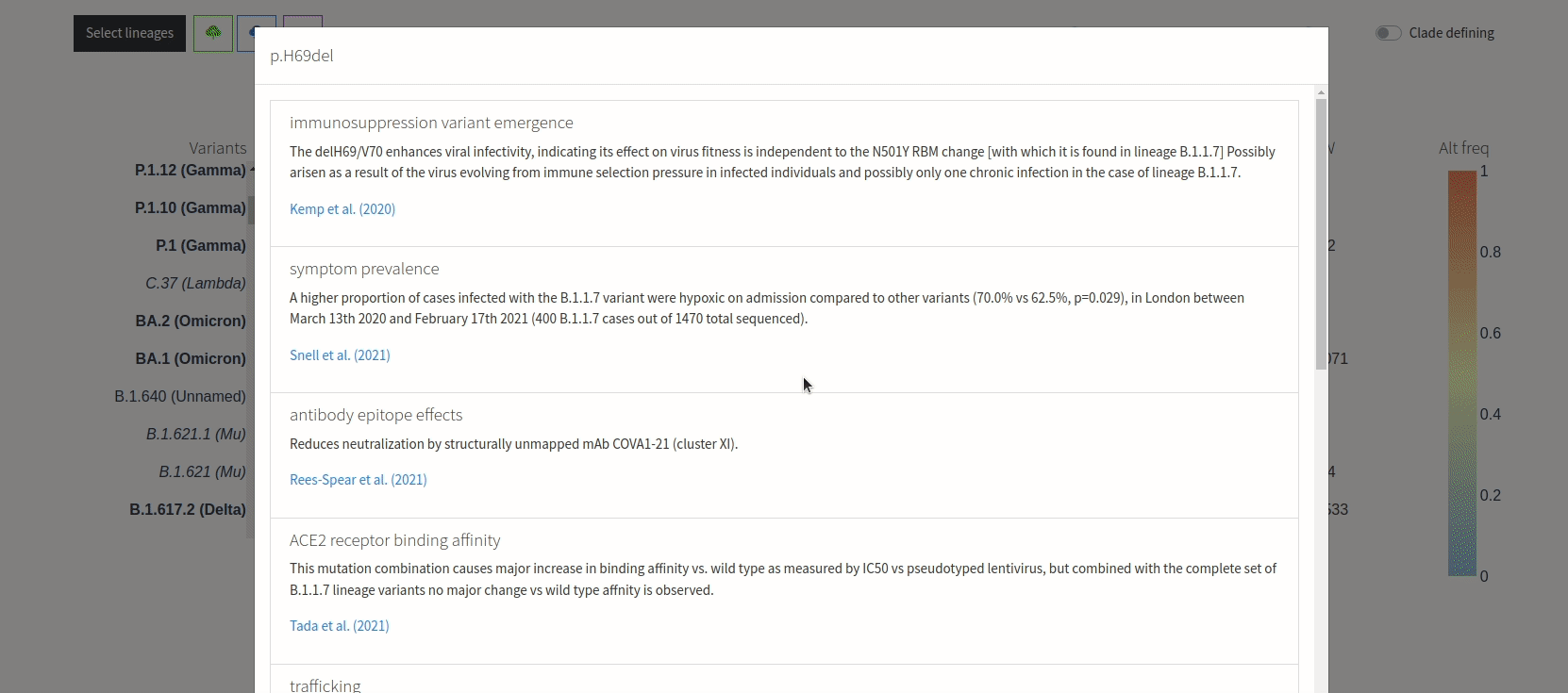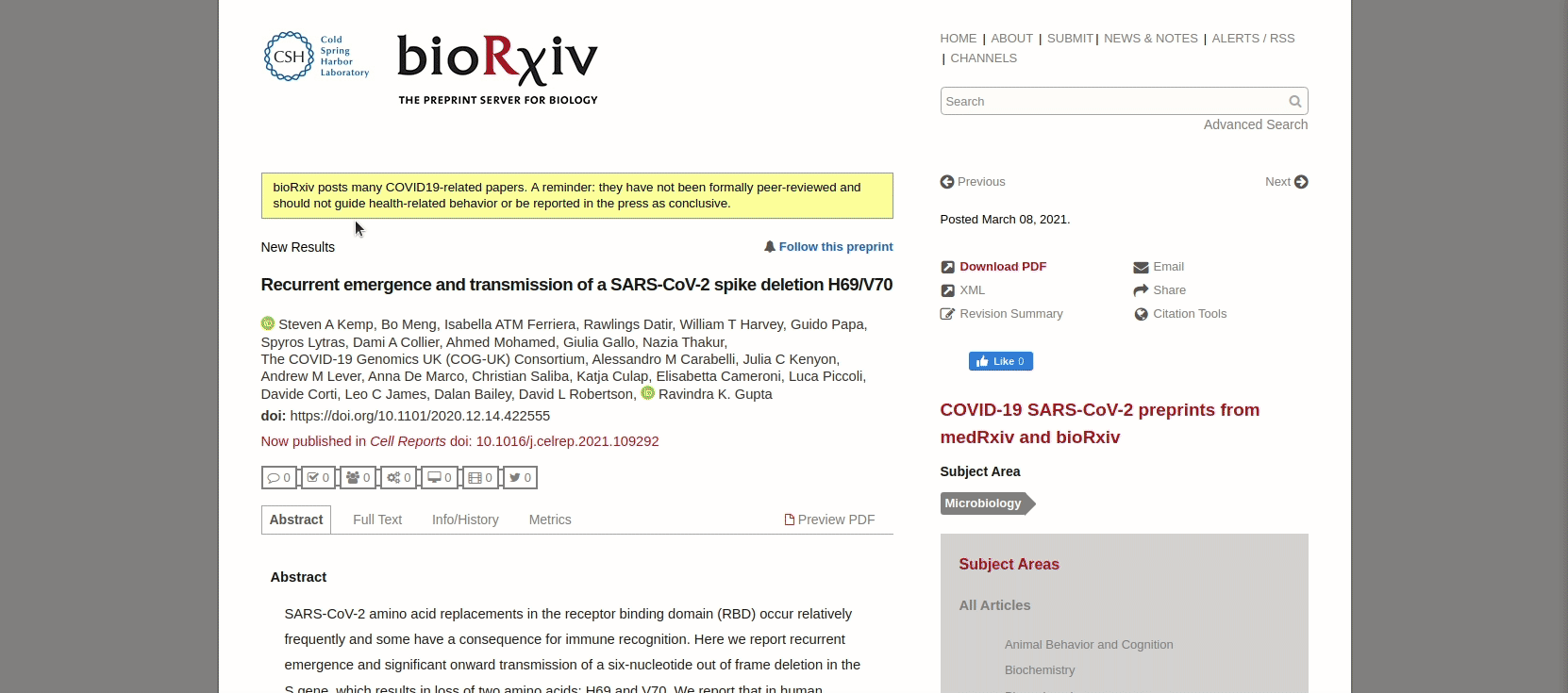
Figure 3. VIRUS-MVP web interface with SARS-CoV-2 sequences grouped into lineages. The central heatmap encodes mutation frequency with functional information available on hover. The bottom histogram shows the distribution of mutations across the genome.
Expanding to other priority pathogens, including H5N1
The flexibility of VIRUS-MVP makes it a powerful tool for monitoring emerging threats beyond SARS-CoV-2 and mpox. The ability to incorporate diverse data types enhances the adaptability of VIRUS-MVP, allowing researchers to expand its use across different surveillance applications. This adaptability is especially crucial for tracking high-risk pathogens like H5N1 avian influenza, where early detection in both clinical and environmental samples, including wastewater, can provide valuable insights before widespread outbreaks occur.
This post was contributed by a Nextflow Ambassador. Ambassadors are passionate individuals who support the Nextflow community. Interested in becoming an ambassador? Read more about it here.
