Nextflowomics: Untangling Biology with Data Analysis Pipelines
The word omics refers to the high throughput measurement of particular biological molecules. For example, genomics is the study of all genetic information of an organism whereas proteomics involves investigating the entire set of proteins produced or modified by an organism. While many associate Nextflow with genomics, there are other disciplines in life sciences that can benefit from scalable data analysis pipelines. In this article, we introduce a selection of omics disciplines and explain how Nextflow pipelines and cloud computing are playing a valuable role in advancing research.
For those wishing to dazzle their friends with one’s mastery of the omics buzzwords, here are some terms you can toss out during your next dinner party:
- Epigenomics – the study of the supporting structure of the genome
- Transcriptomics – the study of RNA molecules (mRNA, rRNA, tRNA) produced in cells
- Proteomics – the large-scale study of proteins
- Metabolomics – the study of chemical processes involving metabolites
- Immunoproteomics – the study of proteins involved in immune response\
- Metagenomics – the study of genetic materials recovered from environmental samples
- Phenomics – the systematic study of phenotypes
There are many more omics where these came from, including glycomics, lipidomics, foodomics…You get the drift.
Multi-omics
Increasingly, multiple omics disciplines are being combined to study biological processes in a more comprehensive manner. As illustrated below, multi-omics techniques are used in everything from personalized medicine and dosing control to discovering new molecular pathways as well as population-level studies.i
Multi-omics has become particularly important in areas such as cancer research, where researchers investigate patient subgroups and biological features across multiple fields to understand the genesis and progression of the disease. With a better understanding of the biology, researchers can design more effective predictive models, develop new clinical treatments, and validate new therapies and drugs.ii
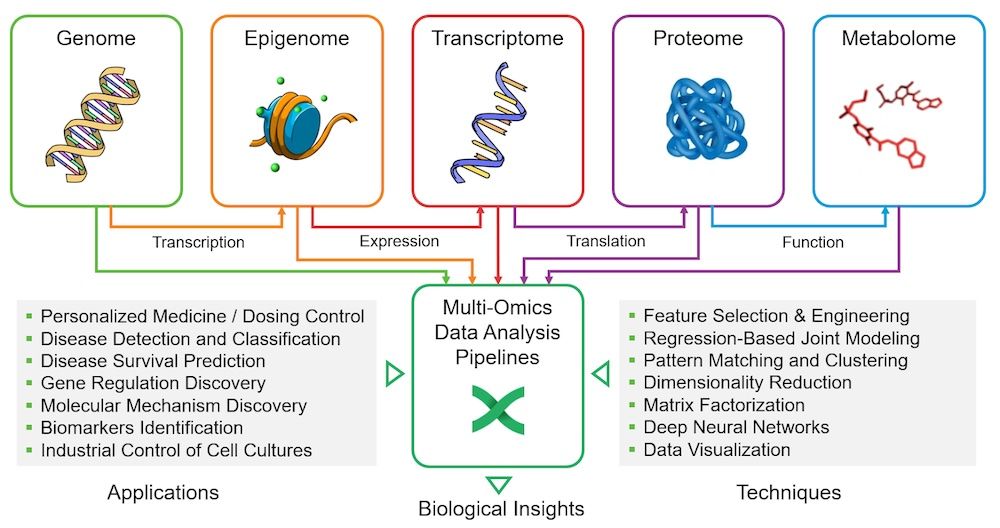
Workflow languages such as Nextflow help researchers to construct and manage data pipelines that parse, process, and analyze vast datasets across the omics disciplines.
Various groups have curated dozens of datasets to explore multi-omics data systematically. Examples include MOPED, the Pancreatic Expression Database, ZikaVR, and ProteomicsDB. However, integrating these vast datasets and studying them in combination is not an easy task. To be effective, pipelines increasingly need to extract and handle data from these public repositories programmatically.
An Open-Air Laboratory of Multi-omics Pipelines
The nf-core project is a showcase for the innovative use of Nextflow pipelines across multiple omics disciplines. A quick survey of curated Nextflow pipelines and datasets available from nf-core shows adoption in the field of genomics and across the omics spectrum. Some examples include:
- Transcriptomics: nf-core/rnaseq – RNA sequencing analysis pipeline using STAR, RSEM, HISAT2 or Salmon with gene/isoform counts and extensive quality control.
- Metagenomics: nf-core/mag – a bioinformatics best-practice pipeline for the assembly, binning, and annotation of metagenomes.
- Metagenomics, pathogen genomics: nf-core/eager – a reproducible state-of-the-art pipeline for the analysts of ancient DNA.
- Metabolomics: nfcore/metaboigniter – a pipeline for pre-processing of mass spectrometry-based metabolomics data with quantification and identification.
- Proteomics: nf-core/proteomicslfq – Proteomics label-free quantification (LFQ) analysis pipeline.
- Proteomics, diaproteomics: nf-core/diaproteomics – an automated quantitative analysis pipeline of DIA proteomics mass spectrometry measurements.
- Proteomics, proteogenomics: nf-core/quantms – a quantitative mass spectrometry workflow supporting proteomics experiments for DDA-LFQ, DDA-Isobaric, and DIA-LFQ quantification.
- Proteomics, peptidomics, immunipeptidomics: nf-core/mhcquant – a bioinformatics pipeline used for the quantitative processing of data-dependent peptidomics data.
- Multi-omics: nf-core/hicar – a pipeline for HiCAR data, a multi-omic co-assay for simultaneous measurement of transcriptome, chromatin accessibility, and cis-regulatory chromatin contacts.
There are numerous additional public pipelines in the Nextflow ecosystem under active development in other fields. Besides chaining together bioinformatics tools, Nextflow is also widely used for other applications. These include image processing and analysis (nf-core/imcyto), statistical modeling, training sets for ML models, and extracting and publishing data to and from public data repositories.
Multi-omics Pipelines in the Cloud
While not every pipeline requires large, scalable compute environments, most omics analysis typically involves the processing of hundreds of input files, each several gigabytes in size.
During the pipeline execution, the intermediate files required for the analysis can expand up to 30x the size of the input data, generating terabytes of data. For multi-omics analysis at scale, cloud computing plays an increasingly critical role. While on-premises HPC environments are widely used in multi-omics analysis, the sheer scale of compute and data requirements often makes the cloud an attractive alternative.
Easy access to the cloud provides many advantages for bioinformaticians. They can:
- Reduce costly data movement requirements when dealing with cloud-resident public datasets by “moving the compute to the data.”
- Access specialized compute resources often unavailable on-premises to support the latest bioinformatics toolsets.
- Easily access specialized and more performant storage solutions to handle analysis across large datasets efficiently.
- Scale quickly, improve throughput, and avoid the management headaches associated with on-premises infrastructure.
With features such as containerized execution, integrations with source-code management systems, sophisticated data handling, and seamless cloud integrations, Nextflow and Nextflow Tower are ideal platforms for multi-omics analysis.
To explore these multi-omics pipelines and start running pipelines in the cloud for free, visit http://tower.nf or contact us below to request a demo.
i Illustration inspired by Multi-Omics Data Factor Analysis by Alex Gurbych, AI – Precision Medicine - April 2021
ii Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes
