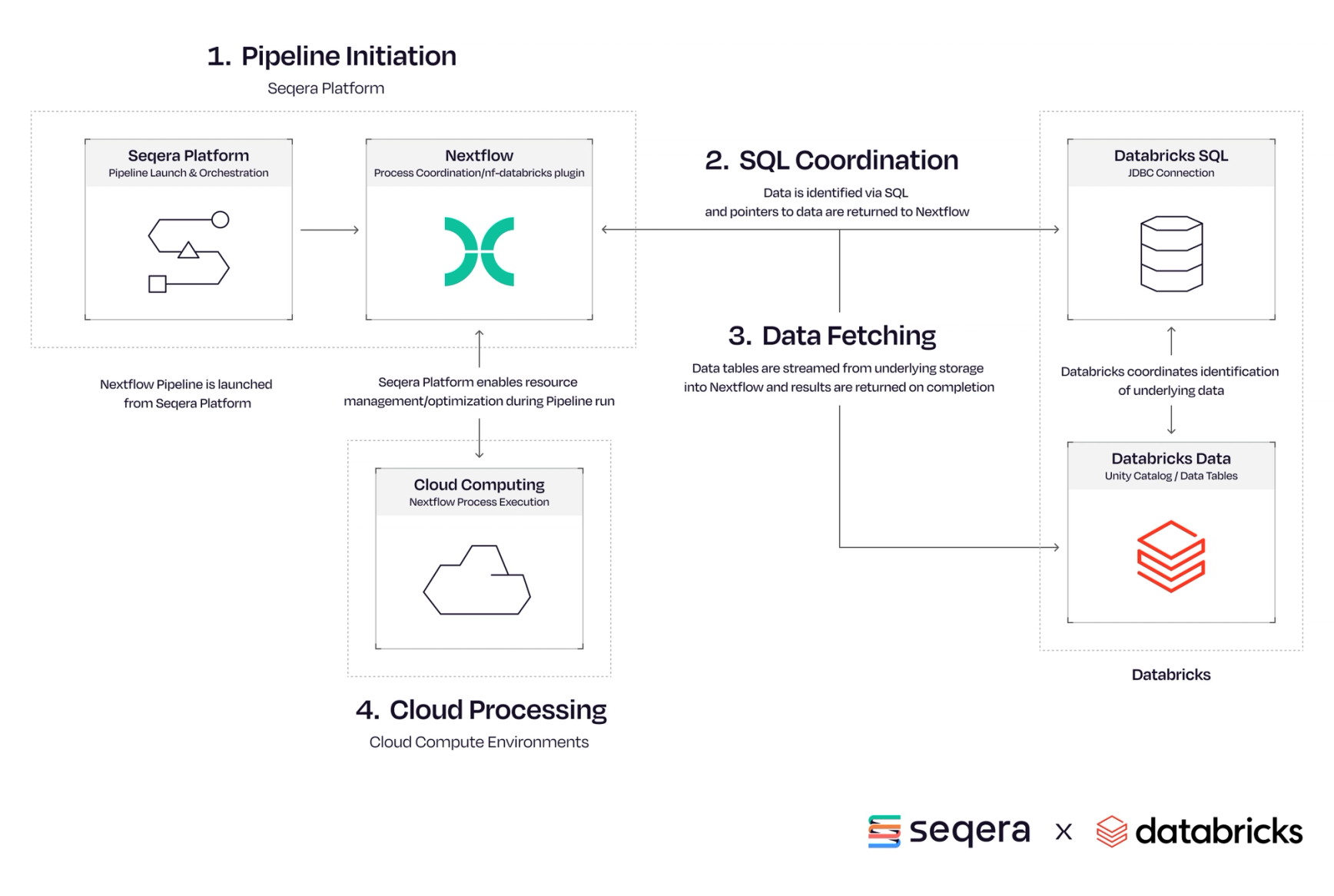
We’re thrilled to announce a powerful new capability for bioinformaticians: native Databricks SQL support in Nextflow via the nf-databricks plugin. This integration bridges the gap between Nextflow’s scalable, reproducible scientific workflows and Databricks’ powerful data analytics platform, enabling end-to-end workflows that are more connected, efficient, and scalable.
Whether you’re preprocessing metadata, querying enterprise-scale data lakes, or storing results for interactive exploration, this enhancement opens exciting possibilities for integrating SQL-driven analytics directly into your Nextflow workflows.
What’s New and Why It Matters
We just introduced a new nf-databricks plugin that brings support for Databricks SQL, further extending Nextflow’s growing ecosystem of database integrations. Using this plugin, users can now connect to Databricks’ analytics engine directly in Nextflow. This update brings several key capabilities:
- →Native Databricks SQL support, eliminating the need for custom workarounds
- →Secure JDBC connectivity for accessing cloud-hosted analytics environments
- →Full SQL operation support, including
fromQuery,sqlExecute, andsqlInsert
This functionality is particularly valuable in bioinformatics and genomics, where working with large, distributed datasets is common. Researchers can now query, transform, and manage data within Databricks directly from their Nextflow pipelines, unlocking key combinations.
- →High-throughput sample tracking and metadata management
- →Multi-omics data integration and cross-platform harmonization
- →Integrated compliance and audit trails for clinical genomics workflows
- →Simplified population-scale variant analysis with dynamic cohort selection
This plugin incorporates and expands the Nextflow’s SQL capabilities provided by nf-sqldb plugin with the support for Databricks specific settings and data sources. The addition of Databricks support continues this trajectory, reflecting our commitment to flexibility and interoperability—empowering users to work across platforms and infrastructures as their projects demand, while maintaining the reproducibility, scalability, and portability that Nextflow is known for.
How to get started with Databricks in Nextflow
Prerequisites before getting started:
• Nextflow 24.04.0 or later
• Java 17 or later
• A Databricks workspace with SQL with a Databricks compute resource
Configuration
In your nextflow.config, define the plugin and Databricks connection:
The JDBC URL follows Databricks' standard connection format. You can find these connection details in your Databricks SQL workspace under the "Connection Details" section. For the authentication, you'll use a Databricks personal access token.
Querying and transforming data
Start with a simple query:
Go further by creating and processing tables:
Real-World Use Case: Genomics Workflow
This workflow demonstrates a complete data processing cycle that's particularly relevant for bioinformatics pipelines. Let’s walk through what's happening here:
Step 1: Data Ingestion from Databricks
The fromQuery operator is fetching pending genomic samples from your Databricks catalog. What's interesting here is that it's creating a Nextflow channel that emits tuples for each row returned by the query - essentially turning SQL results into streamable data that can flow through your pipeline.
Step 2: Data Transformation
This is where Nextflow's functional approach shines. The code is transforming the raw database rows into a format suitable for processing:
- →
row[0]- the sample ID - →
file(row[1])- converting the file path string into a Nextflow file object - →
row[2]- the parameters
Step 3: Analysis and Result Storage
The mysterious ANALYZE_SAMPLES process (not shown in the snippet) does the actual genomic analysis. Then comes the clever part:
This pushes the analysis results back into Databricks. But notice how it doesn't stop there...
Step 4: Status Update Pattern
This final step updates the original records to mark them as completed. The use of parameterized SQL (? placeholders) is a nice touch for safety and flexibility.
Technical Insights
What makes this pattern particularly powerful for bioinformatics workflows:
- State management: This approach maintains processing state in Databricks, enabling better tracking of large-scale genomic analyses, outside of a single Nextflow run.
- Unity catalog Integration: This uses Databricks' Unity Catalog (notice the
ap_demo.nextflowtestnamespace pattern in the examples), which provides governance and tracking capabilities. - Scalable processing: By querying only pending samples, you can run this workflow repeatedly without reprocessing completed work - essential for large genomic datasets.
This hybrid approach addresses a common pain point: how do you track sample processing status across multiple pipeline runs while maintaining Nextflow's reproducibility guarantees?
The clever bit here is that you're using Databricks as both:
- →A metadata store (tracking what needs processing)
- →A results repository (storing outputs and metrics)
This pattern would be particularly useful for clinical genomics pipelines where you need audit trails and processing history.
Technical Notes and Limitations
- →Plugin version: nf-databricks@0.1.0
- →JDBC driver: Official Databricks JDBC driver
- →Network access: Ensure Databricks endpoints are reachable from your execution environment
For JDBC configuration details, refer to Databricks JDBC OSS Docs.
Join the Community
This integration was developed with contributions from the community, including PR #25 and PR #28. We're eager to hear how you're using it!
- →Report issues or request features on the nf-databricks GitHub repo
- →Join conversations in the Nextflow community channels
Final Thoughts
The integration of Databricks SQL into Nextflow represents a significant leap forward in workflow-enabled data analytics. By combining Databricks’ scalable compute power with Nextflow’s flexibility and reproducibility, we’re enabling scientific teams to innovate faster and with greater confidence.
Give it a try—and let us know what you build!
Stay up-to-date with the Seqera Newsletter

