Breakthrough performance and cost-efficiency with the new Fusion file system
Introduction
Running data-intensive pipelines at scale can be complex, particularly when handling the distributed file storage and data transfers required by such applications. This is particularly true for genomics pipelines which tend to involve large datasets. Individual tasks can easily process hundreds of GBs of data and generate similarly large outputs. These large, data-intensive workloads are ill-suited to cloud environments commonly optimized for stateless, shared-nothing computation.
This post discusses the pros and cons of different data-sharing architectures for Nextflow pipelines. It explains how organizations can boost pipeline throughput, reduce costs, and simplify management with Seqera Lab’s Fusion 2.0 file system. We will show how Fusion can help to:
- Improve pipeline throughput by up to 2.2x;
- Deliver performance similar to Amazon FSx for Lustre, without the complexity, and at much lower cost;
- Reduce pipeline run costs including long-term storage by up to 76%.
Data sharing architectures - considering the pros and cons
Nextflow mitigates some of the problems described above by enabling popular object storage services such as AWS S3 to serve as a shared data store during pipeline execution. Having Nextflow tasks access shared S3 buckets directly offers several advantages: S3 is easy to deploy, relatively inexpensive, highly scalable, and supports configurable lifecycle rules that can expire old files automatically to simplify administration and reduce costs.
Despite S3’s obvious advantages, there are challenges, however. Some of these issues include:
- Compute and network overhead: when S3 is used for shared storage, each task needs to copy data from an S3 bucket to a local file system, process the data, and copy results back to the bucket. Pipelines often involve thousands of tasks. These sequential steps of reading, processing, and writing data for every task translate into additional compute requirements, reduced throughput, and network congestion affecting pipeline reliability;
- Per-instance storage: When copying large files from S3 to machine instances, users need to be concerned about available scratch storage on the instance. Since multiple containers typically share a VM, the ephemeral storage associated with VMs is typically not sufficient — it’s not uncommon to require a TB or more of locally attached Elastic Block Storage (EBS), adding cost and complexity to the compute environment. Also, users need to worry about sizing per-instance storage for their workloads;
- Additional utilities: Finally, to facilitate movement to and from S3 buckets, access to the
aws s3 cpcommand (or s3 API) is required. Organizations typically need to use an Amazon Machine Image (AMI) that has the AWS CLI tools pre-installed or provide the AWS CLI in their containers, adding complexity for pipeline developers.
Another approach is to use a shared file system such as NFS or cloud-native file systems such as Amazon EFS or Amazon FSx for Lustre to share data between tasks. By presenting a POSIX file system interface, tasks can access data on the shared file system directly, avoiding the overhead of copying data back and forth from S3 for every task. This makes pipelines more reliable and helps deliver better performance under load. Users can also avoid pre-installing the AWS CLI in AMIs or containers. However, while shared file systems solve some problems, they bring other challenges:
- Cost and complexity: Unlike S3, provisioning and managing shared file systems can be difficult. Users must deploy file systems in advance, configure VPCs appropriately, ensure that file systems are mounted by each instance, and map file system mount points to containers. File systems are also more expensive than S3 storage;
- File system scaling: Depending on the file system, you may need to manage capacity yourself. For example, while AWS EFS can autoscale, changing the size of an FSx for Lustre file system involves modifying the file system via the FSx for Lustre CLI or API, during which time it is unavailable. This means that administrators tend to oversize their file systems to avoid running out of space, adding costs;
- File system permissions: While POSIX file systems are convenient, users need to worry about details such as user ids, group ids, and Linux file system permissions to ensure that files written by one containerized task can be read by another;
- Local scratch storage: The use of a shared file system doesn’t necessarily avoid the need for local instance storage. Tasks may read data from the shared file system but still process it locally on scratch storage before copying results back to the shared file system. This means that data is still frequently replicated and copied, and users may need to deploy block storage with their machine instances;
- Staging data from S3: Similarly, using a file system doesn’t always avoid the need to copy data either. Genomics datasets tend to be stored on inexpensive object storage. While data doesn’t need to be shuttled to and from S3 for every task, data is typically staged at the beginning of pipeline execution. Results are typically copied back to S3 for archival use after the pipeline runs. While this sounds easy enough, it is worth remembering that before a single byte can be written to the shared file system, users will need to spin up a VM and mount the file system to facilitate this data transfer, adding complexity to the overall workflow.
The bottom line is that most shared file systems were designed for a pre-cloud era and meant to run on static HPC clusters with fast, switched network interconnects. By contrast, cloud environments are characterized by ephemeral clusters, dynamic scaling, and limited bandwidth. Clearly, none of the approaches described above are ideal.
Introducing Fusion file system
Recognizing the many challenges associated with managing data in distributed pipelines, we decided to work on finding a better solution.
We started from the assumption that object storage services such as AWS S3 are the ideal solution for cloud native applications because they are cheap, fast, infinitely scalable and are easy to manage. The only gap was that S3 is not compatible with most data analysis tools that generally rely on a POSIX file system interface.
To solve this problem, we developed the Fusion file system. Fusion file system aims to bridge the gap between cloud-native storage and data analysis workflows by implementing a thin client that allows pipeline jobs to access object storage using a standard POSIX interface, thus simplifying and speeding up most operations.
From a technical perspective, this solution is implemented as a FUSE driver, a well known system interface that runs in the user space (i.e., meaning that it does not require administrative privileges to be configured). As readers may be aware, there are several open-source software (OSS) drivers that allow cloud buckets to be mounted as a local file system. Among these are s3fs, gcs-fuse and azure-blob-fuse.
Indeed, in our initial efforts to solve this problem (Fusion 1.0) we looked to these open-source projects, but quickly ran into problems. To solve these, we decided to develop our own solution from scratch (Fusion 2.0). We designed the Fusion file system specifically for Nextflow data analysis pipelines, to take advantage of Nextflow task segregation and its idempotent execution model.
Our improved Fusion driver differs from other OSS solutions in that it is mounted in the container where Nextflow tasks execute and has a minimal memory and CPU usage footprint.
The Fusion file system driver has the following advantages over alternative FUSE driver implementations:
- Fusion file system implements a lazy download and upload algorithm that runs in the background to transfer files in parallel to and from S3 in a non-sequential manner for maximum performance;
- It makes aggressive use of caching and takes advantage of NVMe storage when available, avoiding the need for additional local scratch storage;
- Fusion implements a non-blocking write so that write performance is not constrained by available network bandwidth to S3 bucket;
- A predictive loading algorithm in Fusion file system makes files available to Nextflow tasks as quickly as possible;
- Zero configuration — Fusion is automatically provisioned by the Wave container service in your Nextflow pipeline. It does not require any manual installation, change in your existing workflow code, or container images.
These features combine to help optimize job execution, improve efficiency, and minimize the time required for transferring input and output data leading to faster more cost-effective analysis pipelines.
Benchmarking Fusion
To validate our solution, we ran a benchmark comparing Fusion with other file-sharing approaches using the RNAseq nf-core pipeline configured with the "test_full" launching profile which uses real world execution dataset.
The pipeline execution was carried out in tower.nf with Nextflow version 22.10.6 using 5 different compute environment configurations:
- Using an S3 bucket as the pipeline work directory (Nextflow standard behavior for AWS cloud);
- Using FSx as the pipeline work directory and setting
process.scratch=truein the Nextflow configuration file (default); - Using FSx as the pipeline work directory and setting
process.scratch=falsein the Nextflow configuration file; - Using Fusion 2.0 as the pipeline work directory and setting
process.scratch=truein the Nextflow configuration file; - Using Fusion 2.0 as the pipeline work directory and setting
process.scratch=falsein the Nextflow configuration file (this configuration requires the use of NVMe SSD instance storage to be effective).
The benchmark was performed on AWS Batch using the EU Ireland region over three availability zones (AZs) — eu-west-1a, eu-west-1b, eu-west-1c. The AWS Batch compute environment was configured using the default ECS Linux 2 AMI, max 500 vCPUs and the BEST_FIT_PROGRESSIVE allocation strategy (default). To minimize execution time variability due to spot instance interruptions.
The AWS Batch compute environments were configured with the instance types c6i, m6i, and r6i. The compute environment using Fusion 2.0 in scratch=false mode was configured with instance types c6id, m6id, and r6id (similar family to the previous ones, providing however NVMe ephemeral storage).
Moreover, compute nodes were configured with an EBS volume of 50 GB (3000 iops, 125 Mb/s throughput) used as scratch storage for the AWS Batch compute environments (1) and (4) described above.
Finally, the FSx for Lustre file system was provisioned in the same EU Ireland region using the scratch, SDD deployment mode (200 MB/s/TiB) with no data compression.
For each pipeline and data-sharing configuration, we captured the following results:
- Wall-time (h): The total execution wall-time (in hours) for the pipeline: the elapsed time from pipeline submission to completion;
- CPU-hours (billable): The total billable CPU-hours for the pipeline calculated as the sum up of each task execution time multiplied by the number of vCPUs used by each task;
- CPU-hours (excluding xfers): The total CPU-hours consumed to carry out the pipeline task and excluding data transfers (xfers). Data transfer time is the time required to download task input data and upload task output data;
- Data xfer time: The total CPU-hours spent transferring data: This is the time spent transferring input and output data from/to S3.
Based on these results, we calculated three figures:
- Efficiency-gain (CPU-hours): The improvement in pipeline CPU-usage efficiency (based on billable CPU-hours) compared to the baseline Amazon S3 method;
- Compute cost reduction (%): Comparing billable CPU-hours for each storage method to the baseline Amazon S3 scenario. This is another way of representing the efficiency gain in CPU-hours;
- Data handling efficiency (%): Measuring the proportion of time spent doing useful work vs. managing data transfers.
The pipeline was executed three times for each compute environment tested. The average result for each pipeline and data-sharing approach for the RNAseq pipeline tested is shown in the following table:
| mode | Wall time (h) | CPU-hours (billable) | CPU-hours (excluding xfers) | data xfer time (hours) | Efficiency gain (CPU-hours) | Compute time reduction (%) | Data handling efficiency (%) |
|---|---|---|---|---|---|---|---|
| Amazon S3 | 11.25 | 766.63 | 545.23 | 221.39 | 1.00 | - | 71.1% |
| FSx for Lustre scratch=true | 7.19 | 406.38 | 378.30 | 28.08 | 1.89x | 47% | 93.1% |
| FSx for Lustre scratch=false | 6.86 | 358.97 | 358.55 | 0.42 | 2.14x | 53% | 99.9% |
| Fusion 2.0 scratch=true | 9.27 | 722.72 | 657.07 | 65.65 | 1.06x | 6% | 90.9% |
| Fusion 2.0 scratch=false (NVMe) | 4.66 | 346.10 | 342.91 | 3.18 | 2.22x | 55% | 99.1% |
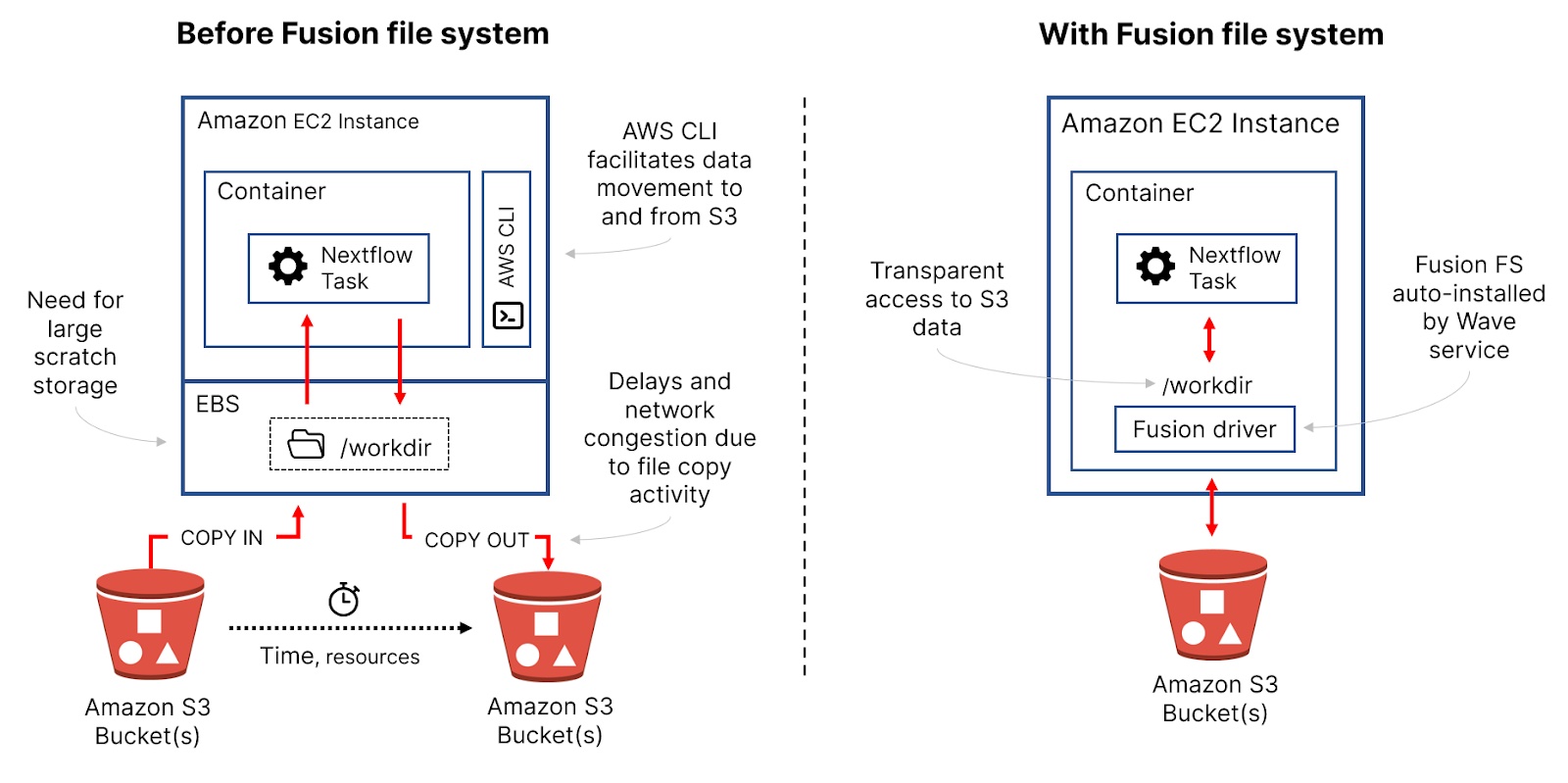
The chart below shows the relative improvement in CPU-usage efficiency based on billable CPU-hours consumed for each storage method compared to the baseline Amazon S3 run. By reducing total CPU hours, we are essentially “speeding up” the workflow. The relative improvements in CPU-hours consumed contribute to the faster pipeline runtimes shown in the wall-time column.
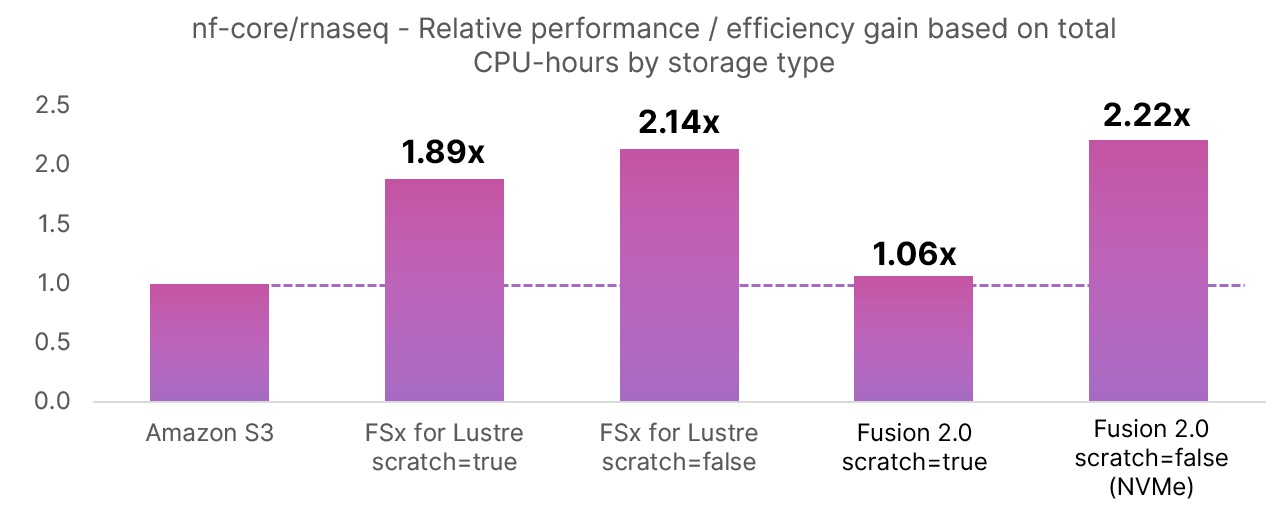
The second chart shows the relative data handling efficiency in percentage terms for each storage configuration tested. This reflects the CPU-hours spent doing useful work as opposed to shuttling data back and forth. Note that Fusion’s data handling efficiency is roughly on par with FSx for Lustre. The standard Amazon S3 method is relatively inefficient (71.1%) because data needs to be copied from and to S3 buckets for every task.
Results
Not surprisingly, using S3 as a shared work directory yielded the slowest result. It required 766.63 billable hours to complete, of which ~221 hours were spent transferring data. We used this result as a baseline for comparing with the other methods.
Using FSx for Lustre, pipeline execution was considerably faster. When scratch mode was enabled, Nextflow created a temporary work directory in the node local disk to carry out the task execution and created symbolic links to reference the task input files. This meant input data was read directly from the shared file system. However, task output data needed to be copied from the local scratch disk to the target work directory in the shared FSx file system. With this setting, the pipeline required 406.38 hours to complete and only ~28 hours was spent transferring data.
By setting scratch mode to false in the FSx scenario, the task used the shared file system path both for reading and writing data. This means that Nextflow did not need extra steps to copy the data output by each task. This is not considered a best practice in the HPC world, however FSx behaves surprisingly well in this scenario, outperforming the default scratch=true mode. In this case, the pipeline required 358.97 hours to complete and only ~0.4 hours was spent transferring data.
It's important to highlight that when using FSx with pipeline input data hosted on S3, the Nextflow main job needs to copy inputs from the S3 bucket(s) into the FSx shared file system prior to execution. This benchmark didn't take this initial data transfer into consideration.
Finally, when using Fusion 2.0 with scratch=true, we observed a substantial reduction in CPU-hours required for data transfer — only ~65.6 hours compared to the ~221.4 hours required when using S3. This resulted in a substantial reduction in billable CPU-hours, even though FSx using the same scratch=true setting performed better.
When the Fusion 2.0 files system was tested with scratch=false, we saw Fusion reach its full potential, we measured an overall execution time of 346.10 billable CPU-hours — a result that is faster than FSx with the same Nextflow scratch setting!
This exceptional performance is possible because Fusion takes advantage of the node’s local NVMe storage for caching purposes and to speed up most disk operations. When using this configuration, pipeline jobs do not need to explicitly transfer data from and to the object storage. The task can access the object storage and read and write directly as if it were a local file system. However Fusion uses the local NVMe storage behind the scenes for I/O disk operations and to transparently download and upload data on-demand from and to the shared S3 bucket.
Impact on pipeline execution and storage costs
The costs of any given run can be broken into two components — one-off costs incurred during each run including EC2 instances, EBS volumes, or network traffic, and ongoing costs such as persistent data storage. We used cost data from actual pipeline runs to model a scenario where data (including final output and intermediate data required for workflow checkpoint/resume functionality) was retained for 30 days.
The use of FSx for Lustre reduces the overall compute time compared to S3, but is more expensive for transfers between availability zones, even within the same region. FSx is much more expensive than S3 for longer-term data retention. Fusion 2.0 associated with the use of NVMe instance storage reduces one-off costs dramatically by minimizing compute time, and it also reduces ongoing costs by using cost-effective S3 storage.
| Configuration | Run costs (one-off) | Data storage per month (ongoing) | Total cost after one month |
|---|---|---|---|
Amazon S3 | $99.05 | $24.56 | $123.61 |
FSx for Lustre scratch=true | $109.21 | $203.86 | $313.07 |
FSx for Lustre scratch=false | $115.02 | $177.97 | $292.99 |
Fusion 2.0 scratch=true | $88.28 | $24.56 | $112.84 |
Fusion 2.0 scratch=false (NVMe) | $49.08 | $19.92 | $68.99 |
All prices are in USD. Data storage costs were calculated based on the total size of the working and output directories (averaged over the three runs for each configuration) multiplied by storage costs at the following rates: $0.023 per GB-month for S3 and $0.154 per GB-month for FSx. Also, on-demand instances were used to avoid compute time variability caused by spot instance retirement.
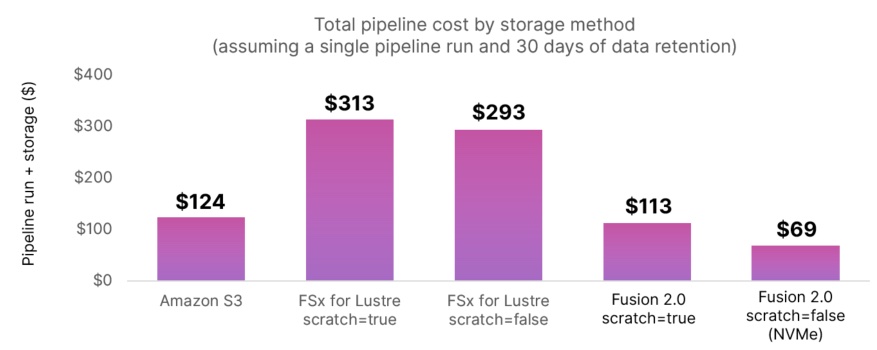 Fusion 2.0 is ~76% less costly than FSx for Lustre and ~44% less than Amazon S3
Fusion 2.0 is ~76% less costly than FSx for Lustre and ~44% less than Amazon S3
Summary
The use of Fusion 2.0 with NVMe storage has a dramatic impact both on both execution time and cost when running common genomics data analysis pipelines. Fusion 2.0 is faster than S3 alone, and up to 76% cheaper when compared to the cost of using FSx for a month. This cost-benefit can be even higher in production scenarios with the use of spot instances.
Other key benefits of Fusion 2.0 when deployed with an NVMe based instance type are:
- No need for additional EBS disks for local scratch;
- Optimal data transfers compared to both the S3 and FSx alternatives;
- No need for Nextflow to manage data transfers between S3 and a shared file system.
Availability and limitations
Fusion 2.0 does not require installation or any change in your pipeline configuration. It only requires the use of a containerized execution environment such as Docker, Kubernetes, or AWS Batch and the use of a S3-compatible file storage.
To use the Fusion file system in your pipeline, add the following snippet to your Nextflow pipeline configuration:
fusion.enabled = true
wave.enabled = trueThese options can be enabled through the web interface when creating the compute environment in Tower cloud.
Presently, Fusion file system is supported by the Nextflow local executor, the AWS Batch executor, Google Cloud Batch, and the Kubernetes executor. Support for Slurm, Grid Engine, and IBM Spectrum LSF batch schedulers has also been added.
Tower Cloud presently only supports Fusion 2.0 with the AWS Batch and the Google Cloud Batch compute environment.
Find more details on available configuration options in the Nextflow documentation.
Future work
In this benchmark, we’ve described the benefits of using Fusion file system over other popular solutions such as Amazon S3 and Amazon FSx for Lustre. We’ve shown that Fusion 2.0, combined with NVMe SSD storage, delivers performance similar to FSx for Lustre but with significantly lower operating and maintenance costs.
Presently, Fusion 2.0 can only be used with S3-compatible object storage and Google Cloud Storage. In the near future, support will be added for Azure Blob Storage. We are also working to improve the performance of Fusion when deployed without the use of local NVMe storage.
Conclusion
Storage is a challenging problem when deploying data pipelines at scale. Existing solutions are either fragile, suboptimal or overly expensive.
With the Fusion file system, we introduce a simple, scalable solution designed to work optimally in cloud-native compute environments. Fusion 2.0 outperforms existing technologies in terms of throughput and data transfer efficiency, and dramatically reduces the costs of operating data pipelines at scale.
To learn more about Fusion file system and Nextflow Tower, contact us or request a demonstration at https://seqera.io/demo/.
Credits
This project took us more than a year of focused research and development. It would not have been possible without the hard work of talented individuals from Seqera. In particular, I want to thank and acknowledge Jordi Deu-Pons who led the Fusion engineering effort. I’d also like to thank Harshil Patel and Rob Syme for their tireless work testing and benchmarking Fusion to achieve these results, and Gordon Sissons for his input and review.
Updates
- On 4th April 2023, this article was updated to reflect the addition of support for Google Cloud Batch, Slurm, Grid Engine and IBM LSF batch schedulers.
